[English] 日本語
 Yorodumi
Yorodumi- EMDB-2100: Location of the dsRNA-dependent polymerase, VP1, in rotavirus par... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2100 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | |||||||||
 Map data Map data | reconstruction of rotavirus DLP+VP1 (polymerase) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  rotavirus / dsRNA-dependent / rotavirus / dsRNA-dependent /  polymerase polymerase | |||||||||
| Function / homology |  Function and homology information Function and homology information virion component => GO:0044423 / viral genome replication / virion component => GO:0044423 / viral genome replication /  RNA-directed RNA polymerase / RNA-directed RNA polymerase /  RNA-dependent RNA polymerase activity / RNA-dependent RNA polymerase activity /  nucleotide binding / DNA-templated transcription / nucleotide binding / DNA-templated transcription /  RNA binding RNA bindingSimilarity search - Function | |||||||||
| Biological species |  Bovine rotavirus Bovine rotavirus | |||||||||
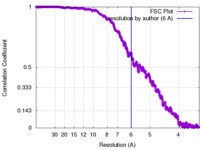
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 6.0 Å cryo EM / Resolution: 6.0 Å | |||||||||
 Authors Authors | Estrozi LF / Settembre EC / Goret G / McClain B / Zhang X / Chen JZ / Grigorieff N / Harrison SC | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2013 Journal: J Mol Biol / Year: 2013Title: Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles. Authors: Leandro F Estrozi / Ethan C Settembre / Gaël Goret / Brian McClain / Xing Zhang / James Z Chen / Nikolaus Grigorieff / Stephen C Harrison /  Abstract: Double-stranded RNA (dsRNA) viruses transcribe and replicate RNA within an assembled, inner capsid particle; only plus-sense mRNA emerges into the intracellular milieu. During infectious entry of a ...Double-stranded RNA (dsRNA) viruses transcribe and replicate RNA within an assembled, inner capsid particle; only plus-sense mRNA emerges into the intracellular milieu. During infectious entry of a rotavirus particle, the outer layer of its three-layer structure dissociates, delivering the inner double-layered particle (DLP) into the cytosol. DLP structures determined by X-ray crystallography and electron cryomicroscopy (cryoEM) show that the RNA coils uniformly into the particle interior, avoiding a "fivefold hub" of more structured density projecting inward from the VP2 shell of the DLP along each of the twelve 5-fold axes. Analysis of the X-ray crystallographic electron density map suggested that principal contributors to the hub are the N-terminal arms of VP2, but reexamination of the cryoEM map has shown that many features come from a molecule of VP1, randomly occupying five equivalent and partly overlapping positions. We confirm here that the electron density in the X-ray map leads to the same conclusion, and we describe the functional implications of the orientation and position of the polymerase. The exit channel for the nascent transcript directs the nascent transcript toward an opening along the 5-fold axis. The template strand enters from within the particle, and the dsRNA product of the initial replication step exits in a direction tangential to the inner surface of the VP2 shell, allowing it to coil optimally within the DLP. The polymerases of reoviruses appear to have similar positions and functional orientations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2100.map.gz emd_2100.map.gz | 7.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2100-v30.xml emd-2100-v30.xml emd-2100.xml emd-2100.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_2100_fsc.xml emd_2100_fsc.xml | 130.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_2100.png emd_2100.png | 207 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2100 http://ftp.pdbj.org/pub/emdb/structures/EMD-2100 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2100 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2100 | HTTPS FTP |
-Related structure data
| Related structure data |  4au6MC  4f5xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2100.map.gz / Format: CCP4 / Size: 20.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2100.map.gz / Format: CCP4 / Size: 20.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | reconstruction of rotavirus DLP+VP1 (polymerase) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.69643 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Rotavirus DLP+VP1
| Entire | Name: Rotavirus DLP+VP1 |
|---|---|
| Components |
|
-Supramolecule #1000: Rotavirus DLP+VP1
| Supramolecule | Name: Rotavirus DLP+VP1 / type: sample / ID: 1000 Oligomeric state: 780 molecules of VP6 form a DLP particle with 12 molecules of VP1, 120 molecules of VP2, 12 molecules of VP3 and 11 dsRNA molecules Number unique components: 5 |
|---|
-Macromolecule #1: Rotavirus polymerase (VP1)
| Macromolecule | Name: Rotavirus polymerase (VP1) / type: protein_or_peptide / ID: 1 / Name.synonym: VP1 Details: The icosahedral 3D reconstruction of rotavirus DLP shows extra-density near the 5-fold axis corresponding to one copy of VP1 attached to the DLP inner surface. Number of copies: 11 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Bovine rotavirus / synonym: Rotavirus Bovine rotavirus / synonym: Rotavirus |
| Molecular weight | Theoretical: 126.326 KDa |
-Macromolecule #2: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 2 / Name.synonym: VP1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Bovine rotavirus / synonym: Rotavirus Bovine rotavirus / synonym: Rotavirus |
-Macromolecule #3: VP2
| Macromolecule | Name: VP2 / type: protein_or_peptide / ID: 3 / Name.synonym: VP2 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Bovine rotavirus / synonym: Rotavirus Bovine rotavirus / synonym: Rotavirus |
-Macromolecule #4: VP3
| Macromolecule | Name: VP3 / type: protein_or_peptide / ID: 4 / Name.synonym: VP3 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Bovine rotavirus / synonym: Rotavirus Bovine rotavirus / synonym: Rotavirus |
-Macromolecule #5: dsRNA
| Macromolecule | Name: dsRNA / type: rna / ID: 5 / Name.synonym: dsRNA / Classification: OTHER / Structure: OTHER / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  Bovine rotavirus / synonym: Rotavirus Bovine rotavirus / synonym: Rotavirus |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Details: Lacy carbon and C-flat |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 30 % / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: Home-made. Vitrification carried out in air at room temperature Method: Blot for 3 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 56540 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 59000 Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder: Eucentric, side-entry / Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Average: 90 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected |
| Date | Jun 1, 2007 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 386 / Average electron dose: 15 e/Å2 / Od range: 1 / Bits/pixel: 8 |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: URO and VEDA |
| Details | Protocol: Rigid body. The Fourier coefficients of VP1 were downweighted by a factor 5 in order to prevent VP1 from being "attracted" by the stronger density of the VP2/6 layer during the fit. matrix 1 0.286272 -0.958148 -0.000156 150.76188 0.573968 0.171358 0.800748 -71.02608 -0.767209 -0.229322 0.599001 -168.57402 matrix 2 -0.084128 -0.546853 -0.832991 184.93534 0.025364 -0.836859 0.546831 15.73129 -0.996132 0.024876 0.084274 -147.45338 matrix 3 0.245240 0.395934 -0.884926 125.30383 -0.558295 -0.688567 -0.462799 80.75011 -0.792568 0.607546 0.052184 -184.30794 matrix 4 0.819204 0.567290 -0.084182 54.27816 -0.370407 0.411304 -0.832843 34.17286 -0.437839 0.713450 0.547070 -228.20418 matrix 5 0.844563 -0.269594 0.462637 70.01368 0.329383 0.942768 -0.051920 -59.62970 -0.422162 0.196234 0.885026 -218.47982 |
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
| Output model |  PDB-4au6: |
 Movie
Movie Controller
Controller