[English] 日本語
 Yorodumi
Yorodumi- EMDB-2074: map of the complex of the HA7 Fab with avian flu virus hemaggluti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2074 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
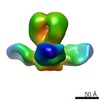
| Title | map of the complex of the HA7 Fab with avian flu virus hemagglutinin by negative staining EM | |||||||||
 Map data Map data | 3D reconstruction from negative staining EM images of the trimeric Fab/hemagglutinin complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HA7 antibody /  Fab / Fab /  hemagglutinin / avian flu virus hemagglutinin / avian flu virus | |||||||||
| Function / homology |  Haemagglutinin, influenzavirus A/B Haemagglutinin, influenzavirus A/B Function and homology information Function and homology information | |||||||||
| Biological species |   Mus musculus (house mouse) / Mus musculus (house mouse) /   unidentified influenza virus unidentified influenza virus | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 18.0 Å negative staining / Resolution: 18.0 Å | |||||||||
 Authors Authors | Du L / Jin L / Zhao G / Sun S / Li J / Li Y / Zheng B / Liddington CL / Zhou Y / Jiang S | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2013 Journal: J Virol / Year: 2013Title: Identification and structural characterization of a broadly neutralizing antibody targeting a novel conserved epitope on the influenza virus H5N1 hemagglutinin. Authors: Lanying Du / Lei Jin / Guangyu Zhao / Shihui Sun / Junfeng Li / Hong Yu / Ye Li / Bo-Jian Zheng / Robert C Liddington / Yusen Zhou / Shibo Jiang /  Abstract: The unabated circulation of the highly pathogenic avian influenza A virus/H5N1 continues to be a serious threat to public health worldwide. Because of the high frequency of naturally occurring ...The unabated circulation of the highly pathogenic avian influenza A virus/H5N1 continues to be a serious threat to public health worldwide. Because of the high frequency of naturally occurring mutations, the emergence of H5N1 variants with high virulence has raised great concerns about the potential transmissibility of the virus in humans. Recent studies have shown that laboratory-mutated or reassortant H5N1 viruses could be efficiently transmitted among mammals, particularly ferrets, the best animal model for humans. Thus, it is critical to establish effective strategies to combat future H5N1 pandemics. In this study, we identified a broadly neutralizing monoclonal antibody (MAb), HA-7, that potently neutralized all tested strains of H5N1 covering clades 0, 1, 2.2, 2.3.4, and 2.3.2.1 and completely protected mice against lethal challenges of H5N1 viruses from clades 1 and 2.3.4. HA-7 specifically targeted the globular head of the H5N1 virus hemagglutinin (HA). Using electron microscopy technology with three-dimensional reconstruction (3D-EM), we discovered that HA-7 bound to a novel and highly conserved conformational epitope that was centered on residues 81 to 83 and 117 to 122 of HA1 (H5 numbering). We further demonstrated that HA-7 inhibited viral entry during postattachment events but not at the receptor-binding step, which is fully consistent with the 3D-EM result. Taken together, we propose that HA-7 could be humanized as an effective passive immunotherapeutic agent for antiviral stockpiling for future influenza pandemics caused by emerging unpredictable H5N1 strains. Our study also provides a sound foundation for the rational design of vaccines capable of inducing broad-spectrum immunity against H5N1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2074.map.gz emd_2074.map.gz | 117.1 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2074-v30.xml emd-2074-v30.xml emd-2074.xml emd-2074.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2074.png EMD-2074.png | 126.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2074 http://ftp.pdbj.org/pub/emdb/structures/EMD-2074 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2074 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2074 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2074.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2074.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction from negative staining EM images of the trimeric Fab/hemagglutinin complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.28 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : the complex of Fab Fragment of HA7 monoclonal antibody and avian ...
| Entire | Name: the complex of Fab Fragment of HA7 monoclonal antibody and avian influenza virus hemagglutinin |
|---|---|
| Components |
|
-Supramolecule #1000: the complex of Fab Fragment of HA7 monoclonal antibody and avian ...
| Supramolecule | Name: the complex of Fab Fragment of HA7 monoclonal antibody and avian influenza virus hemagglutinin type: sample / ID: 1000 / Details: The trimeric sample was monodisperse. Oligomeric state: one trimeric hemagglutinin binds to three Fab Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 370 KDa / Theoretical: 370 KDa / Method: based on protein sequences and SDS-PAGE |
-Macromolecule #1: Fab Fragment of HA7 monoclonal antibody
| Macromolecule | Name: Fab Fragment of HA7 monoclonal antibody / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Oligomeric state: trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) / synonym: House Mouse Mus musculus (house mouse) / synonym: House Mouse |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
-Macromolecule #2: Avian influenza virus hemagglutinin
| Macromolecule | Name: Avian influenza virus hemagglutinin / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Oligomeric state: trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   unidentified influenza virus unidentified influenza virus |
| Sequence | InterPro:  Haemagglutinin, influenzavirus A/B Haemagglutinin, influenzavirus A/B |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 50mM NaCl, 10mM Tris-HCL,1mM CaCl2, pH7.4 |
|---|---|
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein floated on 2% w/v uranyl formate for 30 seconds |
| Grid | Details: 300 mesh copper grid with carbon support, glow discharged in vacuumed air |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 70000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 3.6 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 3.6 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 70,000 times magnification. |
| Date | Aug 7, 2011 |
| Image recording | Category: CCD / Film or detector model: GENERIC TVIPS (4k x 4k) / Number real images: 53 / Average electron dose: 50 e/Å2 / Bits/pixel: 8 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 18.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN2 / Details: C3 symmetry was imposed in refinement / Number images used: 3172 ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 18.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN2 / Details: C3 symmetry was imposed in refinement / Number images used: 3172 |
| Details | Image processing was done using EMAN2.The particles were selected by a semi-automatic selection using e2boxer. |
 Movie
Movie Controller
Controller