[English] 日本語
 Yorodumi
Yorodumi- EMDB-2070: Octameric structure of IMPDH from Pseudomonas aeruginosa in prese... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2070 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
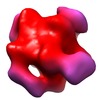
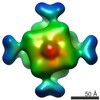
| Title | Octameric structure of IMPDH from Pseudomonas aeruginosa in presence of Mg-ATP | |||||||||
 Map data Map data | Cryo-EM reconstruction of IMPDH from Pseudomonas Aeruginosa in presence of Mg-ATP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | IMPDH /  nucleotide metabolism / CBS domains nucleotide metabolism / CBS domains | |||||||||
| Biological species |   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 16.0 Å cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Labesse G / Vaupre L / alard-Arnaud I / Alexandre T / Lai Kee Him J / Raynal B / Bron P / Munier-Lehmann H | |||||||||
 Citation Citation |  Journal: Structure / Year: 2013 Journal: Structure / Year: 2013Title: MgATP regulates allostery and fiber formation in IMPDHs. Authors: Gilles Labesse / Thomas Alexandre / Laurène Vaupré / Isabelle Salard-Arnaud / Joséphine Lai Kee Him / Bertrand Raynal / Patrick Bron / Hélène Munier-Lehmann /  Abstract: Inosine-5'-monophosphate dehydrogenase (IMPDH) is a rate-limiting enzyme in nucleotide biosynthesis studied as an important therapeutic target and its complex functioning in vivo is still puzzling ...Inosine-5'-monophosphate dehydrogenase (IMPDH) is a rate-limiting enzyme in nucleotide biosynthesis studied as an important therapeutic target and its complex functioning in vivo is still puzzling and debated. Here, we highlight the structural basis for the regulation of IMPDHs by MgATP. Our results demonstrate the essential role of the CBS tandem, conserved among almost all IMPDHs. We found that Pseudomonas aeruginosa IMPDH is an octameric enzyme allosterically regulated by MgATP and showed that this octameric organization is widely conserved in the crystal structures of other IMPDHs. We also demonstrated that human IMPDH1 adopts two types of complementary octamers that can pile up into isolated fibers in the presence of MgATP. The aggregation of such fibers in the autosomal dominant mutant, D226N, could explain the onset of the retinopathy adRP10. Thus, the regulatory CBS modules in IMPDHs are functional and they can either modulate catalysis or macromolecular assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2070.map.gz emd_2070.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2070-v30.xml emd-2070-v30.xml emd-2070.xml emd-2070.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  image2070.png image2070.png | 105.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2070 http://ftp.pdbj.org/pub/emdb/structures/EMD-2070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2070 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2070.map.gz / Format: CCP4 / Size: 12.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2070.map.gz / Format: CCP4 / Size: 12.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of IMPDH from Pseudomonas Aeruginosa in presence of Mg-ATP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : IMPDH from Pseudomonas aeruginosa in presence of Mg-ATP filtered ...
| Entire | Name: IMPDH from Pseudomonas aeruginosa in presence of Mg-ATP filtered at 16A resolution |
|---|---|
| Components |
|
-Supramolecule #1000: IMPDH from Pseudomonas aeruginosa in presence of Mg-ATP filtered ...
| Supramolecule | Name: IMPDH from Pseudomonas aeruginosa in presence of Mg-ATP filtered at 16A resolution type: sample / ID: 1000 / Oligomeric state: octameric / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 430 KDa |
-Macromolecule #1: Inosine-5'-monophosphate dehydrogenase
| Macromolecule | Name: Inosine-5'-monophosphate dehydrogenase / type: protein_or_peptide / ID: 1 / Name.synonym: IMPDH / Number of copies: 8 / Oligomeric state: octamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
| Molecular weight | Theoretical: 430 KDa |
| Recombinant expression | Organism:  Bacteria (eubacteria) Bacteria (eubacteria) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM potassium phosphate pH 8 and 0.1 M KCl |
| Grid | Details: Quantifoil R2.2, glow discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 98 % / Chamber temperature: 102.15 K / Instrument: GATAN CRYOPLUNGE 3 / Method: Blot for 1 second |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 130137 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 100000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 100000 |
| Specialist optics | Energy filter - Name: JEOL / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Sample stage | Specimen holder: 626 Gatan cryo-Holder / Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Min: 93.15 K / Max: 96.15 K / Average: 95.15 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 200,000 times magnification |
| Details | Image acquisition is done directly on the CCD camera. It explains why the nominal magnification is 100000 and calibrated magnification 130137. |
| Date | Nov 1, 2011 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (4k x 4k) / Number real images: 80 / Average electron dose: 18 e/Å2 / Details: Images directly recorded with CCD camera |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D4 (2x4 fold dihedral ) / Resolution.type: BY AUTHOR / Resolution: 16.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IMAGIC, FREALIGN / Number images used: 22577 ) / Resolution.type: BY AUTHOR / Resolution: 16.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IMAGIC, FREALIGN / Number images used: 22577 |
| Details | The particles were selected using a semi-automatic selection program (Boxer). |
 Movie
Movie Controller
Controller