[English] 日本語
 Yorodumi
Yorodumi- EMDB-1617: C12 symmetrised 3D reconstruction of the Shigella flexneri T3SS n... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1617 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | C12 symmetrised 3D reconstruction of the Shigella flexneri T3SS needle complex from negatively stained sample electron micrographs | |||||||||
 Map data Map data | This is a 3D reconstruction of the Shigella flexneri 'needle complex' from negative stain images, done with C12 symmetry imposed. The resolution is 21-25A. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Shigella flexneri Type III secretion system Needle complex Microbial pathogenesis Shigella flexneri Type III secretion system Needle complex Microbial pathogenesis | |||||||||
| Function / homology | Yop virulence translocation protein R / Type III exporter system, secretion apparatus protein BsaZ /  Type III secretion system outer membrane pore YscC/HrcC / Type III secretion system outer membrane pore YscC/HrcC /  Type III secretion system, needle protein / Flagellar M-ring , N-terminal / Type III secretion system, needle protein / Flagellar M-ring , N-terminal /  protein secretion / protein secretion /  : / cell outer membrane / : / cell outer membrane /  protein transport protein transport Function and homology information Function and homology information | |||||||||
| Biological species |   Shigella flexneri (bacteria) Shigella flexneri (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 25.0 Å negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Hodgkinson JL / Horsley A / Stabat D / Simon M / Johnson S / da Fonseca PCA / Morris EP / Wall JS / Lea SM / Blocker AJ | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2009 Journal: Nat Struct Mol Biol / Year: 2009Title: Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Authors: Julie L Hodgkinson / Ashley Horsley / David Stabat / Martha Simon / Steven Johnson / Paula C A da Fonseca / Edward P Morris / Joseph S Wall / Susan M Lea / Ariel J Blocker /  Abstract: Type III secretion systems (T3SSs) mediate bacterial protein translocation into eukaryotic cells, a process essential for virulence of many Gram-negative pathogens. They are composed of a cytoplasmic ...Type III secretion systems (T3SSs) mediate bacterial protein translocation into eukaryotic cells, a process essential for virulence of many Gram-negative pathogens. They are composed of a cytoplasmic secretion machinery and a base that bridges both bacterial membranes, into which a hollow, external needle is embedded. When isolated, the latter two parts are termed the 'needle complex'. An incomplete understanding of the structure of the needle complex has hampered studies of T3SS function. To estimate the stoichiometry of its components, we measured the mass of its subdomains by scanning transmission electron microscopy (STEM). We determined subunit symmetries by analysis of top and side views within negatively stained samples in low-dose transmission electron microscopy (TEM). Application of 12-fold symmetry allowed generation of a 21-25-A resolution, three-dimensional reconstruction of the needle complex base, revealing many new features and permitting tentative docking of the crystal structure of EscJ, an inner membrane component. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1617.map.gz emd_1617.map.gz | 13.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1617-v30.xml emd-1617-v30.xml emd-1617.xml emd-1617.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  1617.gif 1617.gif 1617.png 1617.png | 32.1 KB 327.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1617 http://ftp.pdbj.org/pub/emdb/structures/EMD-1617 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1617 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1617 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1617.map.gz / Format: CCP4 / Size: 22.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1617.map.gz / Format: CCP4 / Size: 22.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a 3D reconstruction of the Shigella flexneri 'needle complex' from negative stain images, done with C12 symmetry imposed. The resolution is 21-25A. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
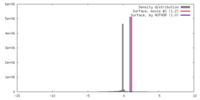
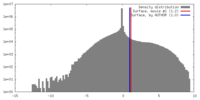
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Shigella flexneri T3SS needle complex
| Entire | Name: Shigella flexneri T3SS needle complex |
|---|---|
| Components |
|
-Supramolecule #1000: Shigella flexneri T3SS needle complex
| Supramolecule | Name: Shigella flexneri T3SS needle complex / type: sample / ID: 1000 Details: Affinity purified using His6 tag on MxiG N-term Contains detergent (see Zenk et al., 2007) Oligomeric state: Large macromolecular complex (total list of components not yet known) Number unique components: 8 |
|---|---|
| Molecular weight | Experimental: 3.6 MDa / Method: STEM analysis |
-Macromolecule #1: Spa24
| Macromolecule | Name: Spa24 / type: protein_or_peptide / ID: 1 / Name.synonym: Spa24 Details: This is the MW of the monomer, which must be cleaved in its cytoplasmic portion during T3SS biogenesis. Oligomeric state: Unknown / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: inner membrane protein Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: inner membrane protein |
| Molecular weight | Theoretical: 240 KDa |
| Sequence | GO:  protein secretion / InterPro: Yop virulence translocation protein R protein secretion / InterPro: Yop virulence translocation protein R |
-Macromolecule #2: Spa40
| Macromolecule | Name: Spa40 / type: protein_or_peptide / ID: 2 / Name.synonym: Spa40 Details: This is the MW of the monomer, which must be cleaved in its cytoplasmic portion during T3SS biogenesis. Oligomeric state: Unknown / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: polytopic-inner membrane protein Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: polytopic-inner membrane protein |
| Molecular weight | Theoretical: 400 KDa |
| Sequence | GO:  protein secretion protein secretionInterPro: Type III exporter system, secretion apparatus protein BsaZ |
-Macromolecule #3: MxiM
| Macromolecule | Name: MxiM / type: protein_or_peptide / ID: 3 / Name.synonym: MxiM / Details: This is the MW of the monomer. / Number of copies: 12 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: outside face of outer membrane Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: outside face of outer membrane |
| Molecular weight | Theoretical: 150 KDa |
| Sequence | GO: cell outer membrane |
-Macromolecule #4: MxiI
| Macromolecule | Name: MxiI / type: protein_or_peptide / ID: 4 / Name.synonym: MxiI / Details: This is the MW of the monomer. / Number of copies: 10 / Oligomeric state: Helical polymer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: periplasmic space Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: periplasmic space |
| Molecular weight | Theoretical: 100 KDa |
| Sequence | GO:  protein transport protein transport |
-Macromolecule #5: MxiG
| Macromolecule | Name: MxiG / type: protein_or_peptide / ID: 5 / Name.synonym: MxiG / Details: This is the MW of the monomer. / Number of copies: 24 / Oligomeric state: Probable homo-24mer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: spans inner membrane Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: spans inner membrane |
| Molecular weight | Theoretical: 430 KDa |
| Sequence | GO:  GO: 0009405 GO: 0009405 |
-Macromolecule #6: MxiJ
| Macromolecule | Name: MxiJ / type: protein_or_peptide / ID: 6 / Name.synonym: MxiJ Details: This is the MW of the monomer with its N-terminal lipid modification site processed (but MW of lipid moiety unknown). Number of copies: 12 / Oligomeric state: Probable homo-dodecamer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: mostly periplasmic face of inner membrane Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: mostly periplasmic face of inner membrane |
| Molecular weight | Theoretical: 250 KDa |
| Sequence | GO:  protein secretion / InterPro: Flagellar M-ring , N-terminal protein secretion / InterPro: Flagellar M-ring , N-terminal |
-Macromolecule #7: MxiH
| Macromolecule | Name: MxiH / type: protein_or_peptide / ID: 7 / Name.synonym: MxiH / Details: This is the MW of the monomer. / Number of copies: 120 / Oligomeric state: helical polymer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: Extracellular Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: Extracellular |
| Molecular weight | Theoretical: 90 KDa |
| Sequence | GO: GO: 0009405 / InterPro:  Type III secretion system, needle protein Type III secretion system, needle protein |
-Macromolecule #8: MxiD
| Macromolecule | Name: MxiD / type: protein_or_peptide / ID: 8 / Name.synonym: MxiD Details: This is the MW of the monomer without it's signal sequence included Number of copies: 12 / Oligomeric state: homo-dodecamer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: Outer membrane Shigella flexneri (bacteria) / Strain: Shigella flexneri M90T / synonym: Dysentery bacillus / Cell: Gram-negative bacterium / Organelle: Type III secretion system / Location in cell: Outer membrane |
| Molecular weight | Theoretical: 620 KDa |
| Sequence | GO:  protein secretion protein secretionInterPro:  Type III secretion system outer membrane pore YscC/HrcC Type III secretion system outer membrane pore YscC/HrcC |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.050 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 10mM Tris pH 8, 0.1% Triton X-100 and 1mM EDTA buffer |
| Staining | Type: NEGATIVE Details: negatively stained in unbuffered 2% aqueous uranyl acetate |
| Grid | Details: 400 mesh copper grids (Athene) covered with holey carbon film over which thin plain carbon was laid were used |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 48600 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 0.9 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 50000 Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 0.9 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: eucentric / Specimen holder model: OTHER |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 100,000 times magnification |
| Details | Low-dose mode was used |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.35 µm / Number real images: 126 / Average electron dose: 20 e/Å2 / Od range: 1 / Bits/pixel: 16 |
- Image processing
Image processing
| Final two d classification | Number classes: 4 |
|---|---|
| Final angle assignment | Details: Imagic beta 87-93 degrees gamma 0-30 degrees |
| Final reconstruction | Applied symmetry - Point group: C12 (12 fold cyclic ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: OTHER / Software - Name: SPIDER IMAGIC5 ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: OTHER / Software - Name: SPIDER IMAGIC5Details: Maps were generated from 41 individual images (selected through image processing out of an initial pool of 3000) Number images used: 41 |
| Details | Numbers of particles given are for final reconstruction. We started with over 3000. Particles were selected using Ximdisp software, cut out to an initial box size of 400 by 400 pixel and coarsened to 2.62A per pixel using Label prior to further processing. |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)