+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1568 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
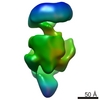
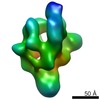
| Title | E.coli core RNA polymerase | |||||||||
 Map data Map data | E.coli RNA polymerase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  RNA polymerase / RNA polymerase /  RNAP / RNAP /  Sigma 54 / Sigma 54 /  transcription initiation / transcription initiation /  sigma factor sigma factor | |||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 23.0 Å cryo EM / Resolution: 23.0 Å | |||||||||
 Authors Authors | Bose D / Pape T / Burrows PC / Rappas M / Wigneshweraraj SR / Buck M / Zhang X | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2008 Journal: Mol Cell / Year: 2008Title: Organization of an activator-bound RNA polymerase holoenzyme. Authors: Daniel Bose / Tillmann Pape / Patricia C Burrows / Mathieu Rappas / Siva R Wigneshweraraj / Martin Buck / Xiaodong Zhang /  Abstract: Transcription initiation involves the conversion from closed promoter complexes, comprising RNA polymerase (RNAP) and double-stranded promoter DNA, to open complexes, in which the enzyme is able to ...Transcription initiation involves the conversion from closed promoter complexes, comprising RNA polymerase (RNAP) and double-stranded promoter DNA, to open complexes, in which the enzyme is able to access the DNA template in a single-stranded form. The complex between bacterial RNAP and its major variant sigma factor sigma(54) remains as a closed complex until ATP hydrolysis-dependent remodeling by activator proteins occurs. This remodeling facilitates DNA melting and allows the transition to the open complex. Here we present cryoelectron microscopy reconstructions of bacterial RNAP in complex with sigma(54) alone, and of RNAP-sigma(54) with an AAA+ activator. Together with photo-crosslinking data that establish the location of promoter DNA within the complexes, we explain why the RNAP-sigma(54) closed complex is unable to access the DNA template and propose how the structural changes induced by activator binding can initiate conformational changes that ultimately result in formation of the open complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1568.map.gz emd_1568.map.gz | 7.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1568-v30.xml emd-1568-v30.xml emd-1568.xml emd-1568.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
| Images |  1568.gif 1568.gif | 31.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1568 http://ftp.pdbj.org/pub/emdb/structures/EMD-1568 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1568 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1568 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1568.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1568.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E.coli RNA polymerase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.6 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of E.coli RNA polymerase and Sigma 54
| Entire | Name: Complex of E.coli RNA polymerase and Sigma 54 |
|---|---|
| Components |
|
-Supramolecule #1000: Complex of E.coli RNA polymerase and Sigma 54
| Supramolecule | Name: Complex of E.coli RNA polymerase and Sigma 54 / type: sample / ID: 1000 / Details: All components are present on SDS-PAGE. / Oligomeric state: monomer of RNAP sigma 54 holoenzyme / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 390 KDa |
-Macromolecule #1: RNA polymerase
| Macromolecule | Name: RNA polymerase / type: protein_or_peptide / ID: 1 / Name.synonym: RNAP / Number of copies: 1 / Oligomeric state: pentamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) / Cell: E.coli Escherichia coli (E. coli) / Cell: E.coli |
| Molecular weight | Theoretical: 390 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) / Recombinant plasmid: pvs10 Escherichia coli (E. coli) / Recombinant plasmid: pvs10 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 10mM Tris-HCl pH8.0, 150mM NaCl, 10mM MgCl2, |
|---|---|
| Grid | Details: Copper 400 mesh |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 83 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG/UT |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 48700 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.1 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 50000 Bright-field microscopy / Cs: 2.1 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Average: 91 K |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Number real images: 100 / Average electron dose: 10 e/Å2 / Details: CCD images |
- Image processing
Image processing
| CTF correction | Details: particles |
|---|---|
| Final two d classification | Number classes: 173 |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: OTHER / Software - Name: Imagic V / Number images used: 4327 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: Situs |
| Details | Protocol: Rigid body. Structure positioned by hand and fitting refined in Situs |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 0.6 |
 Movie
Movie Controller
Controller