[English] 日本語
 Yorodumi
Yorodumi- EMDB-1195: Cryo-electron microscopy studies of human TFIID: conformational b... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1195 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. | |||||||||
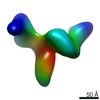
 Map data Map data | Average 3D structure of human TFIID in solution (cryo-EM) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 32.0 Å cryo EM / Resolution: 32.0 Å | |||||||||
 Authors Authors | Grob P / Cruse MJ / Inouye C / Peris M / Penczek PA / Tjian R / Nogales E | |||||||||
 Citation Citation |  Journal: Structure / Year: 2006 Journal: Structure / Year: 2006Title: Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Authors: Patricia Grob / Michael J Cruse / Carla Inouye / Marian Peris / Pawel A Penczek / Robert Tjian / Eva Nogales /  Abstract: The multisubunit transcription factor TFIID is essential for directing eukaryotic promoter recognition and mediating interactions with activators/cofactors during assembly of the preinitiation ...The multisubunit transcription factor TFIID is essential for directing eukaryotic promoter recognition and mediating interactions with activators/cofactors during assembly of the preinitiation complex. Despite its central role in transcription initiation and regulation, structural knowledge of the TFIID complex has so far been largely limited to electron microscopy studies of negatively stained samples. Here, we present a cryo-electron microscopy 3D reconstruction of the large endogenous human TFIID complex. The improved cryopreservation has allowed for a more detailed definition of the structural elements in the complex and for the detection, by an extensive statistical analysis of the data, of a conformational opening and closing of the cavity central to the TFIID architecture. We propose that these density rearrangements in the structure are a likely reflection of the plasticity of the interactions between TFIID and its many partner proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1195.map.gz emd_1195.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1195-v30.xml emd-1195-v30.xml emd-1195.xml emd-1195.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| Images |  1195.gif 1195.gif | 13.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1195 http://ftp.pdbj.org/pub/emdb/structures/EMD-1195 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1195 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1195 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1195.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1195.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Average 3D structure of human TFIID in solution (cryo-EM) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human TFIID
| Entire | Name: human TFIID Transcription factor II D Transcription factor II D |
|---|---|
| Components |
|
-Supramolecule #1000: human TFIID
| Supramolecule | Name: human TFIID / type: sample / ID: 1000 Details: The sample was prepared from a single preparation of TFIID, immunopurified from HeLa cell nuclear extrats (TAF130 antibody) Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 1.0 MDa / Theoretical: 1.0 MDa / Method: Chromatography, sequence |
-Macromolecule #1: TFIID
| Macromolecule | Name: TFIID / type: protein_or_peptide / ID: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: human / Cell: HeLa / Organelle: Nucleus / Location in cell: Nucleus Homo sapiens (human) / synonym: human / Cell: HeLa / Organelle: Nucleus / Location in cell: Nucleus |
| Molecular weight | Experimental: 1.0 MDa / Theoretical: 1.0 MDa |
| Recombinant expression | Organism:  HeLa HeLa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.025 mg/mL |
|---|---|
| Buffer | pH: 7.9 Details: 25mM HEPES, 0.1 mM EDTA, 12.5mM MgCl2, 200mM KCl, 0.03% NP 40 |
| Grid | Details: 400 mesh copper grid with holey carbon |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 93 K / Instrument: OTHER Details: Vitrification instrument: Vitrobot. rince once with sample buffer before blotting |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50200 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 50000 Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: side entry / Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Average: 90 K |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 12.7 µm / Number real images: 90 / Average electron dose: 16 e/Å2 / Bits/pixel: 14 |
- Image processing
Image processing
| CTF correction | Details: whole micrograph |
|---|---|
| Final angle assignment | Details: SPIDER, theta 90 degrees, phi 359.9 |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 32.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Spider, IMAGIC Details: final reconstruction from 8016 single particles, refined by iterative projection matching Number images used: 8016 |
| Details | additional continuous thin carbon layer |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















