[English] 日本語
 Yorodumi
Yorodumi- EMDB-1045: Cryo-EM reveals an active role for aminoacyl-tRNA in the accommod... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1045 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. | |||||||||
 Map data Map data | 70S aa-tRNA-EF-Tu.GDP.kirromycin complex | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationguanyl-nucleotide exchange factor complex /  guanosine tetraphosphate binding / translational elongation / guanosine tetraphosphate binding / translational elongation /  translation elongation factor activity / response to antibiotic / translation elongation factor activity / response to antibiotic /  GTPase activity / GTP binding / GTPase activity / GTP binding /  RNA binding / RNA binding /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli (E. coli) / synthetic construct (others) Escherichia coli (E. coli) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 16.8 Å cryo EM / Resolution: 16.8 Å | |||||||||
 Authors Authors | Valle M / Sengupta J / Swami NK / Grassucci RA / Burkhardt N / Nierhaus KH / Agrawal RK / Frank J | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2002 Journal: EMBO J / Year: 2002Title: Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. Authors: Mikel Valle / Jayati Sengupta / Neil K Swami / Robert A Grassucci / Nils Burkhardt / Knud H Nierhaus / Rajendra K Agrawal / Joachim Frank /  Abstract: During the elongation cycle of protein biosynthesis, the specific amino acid coded for by the mRNA is delivered by a complex that is comprised of the cognate aminoacyl-tRNA, elongation factor Tu and ...During the elongation cycle of protein biosynthesis, the specific amino acid coded for by the mRNA is delivered by a complex that is comprised of the cognate aminoacyl-tRNA, elongation factor Tu and GTP. As this ternary complex binds to the ribosome, the anticodon end of the tRNA reaches the decoding center in the 30S subunit. Here we present the cryo- electron microscopy (EM) study of an Escherichia coli 70S ribosome-bound ternary complex stalled with an antibiotic, kirromycin. In the cryo-EM map the anticodon arm of the tRNA presents a new conformation that appears to facilitate the initial codon-anticodon interaction. Furthermore, the elbow region of the tRNA is seen to contact the GTPase-associated center on the 50S subunit of the ribosome, suggesting an active role of the tRNA in the transmission of the signal prompting the GTP hydrolysis upon codon recognition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1045.map.gz emd_1045.map.gz | 8.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1045-v30.xml emd-1045-v30.xml emd-1045.xml emd-1045.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| Images |  1045.gif 1045.gif | 45.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1045 http://ftp.pdbj.org/pub/emdb/structures/EMD-1045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1045 | HTTPS FTP |
-Related structure data
| Related structure data |  1ls2MC  1lu3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1045.map.gz / Format: CCP4 / Size: 9.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1045.map.gz / Format: CCP4 / Size: 9.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 70S aa-tRNA-EF-Tu.GDP.kirromycin complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.69 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 70S aa-tRNA-EF-Tu.GDP.kirromycin complex from E.coli
| Entire | Name: 70S aa-tRNA-EF-Tu.GDP.kirromycin complex from E.coli |
|---|---|
| Components |
|
-Supramolecule #1000: 70S aa-tRNA-EF-Tu.GDP.kirromycin complex from E.coli
| Supramolecule | Name: 70S aa-tRNA-EF-Tu.GDP.kirromycin complex from E.coli / type: sample / ID: 1000 / Number unique components: 6 |
|---|---|
| Molecular weight | Experimental: 2.5 MDa / Theoretical: 2.5 MDa |
-Supramolecule #1: E.coli 70S ribosome
| Supramolecule | Name: E.coli 70S ribosome / type: complex / ID: 1 / Details: 10 pmol / Recombinant expression: No / Ribosome-details: ribosome-prokaryote: ALL |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: aa-tRNA
| Macromolecule | Name: aa-tRNA / type: rna / ID: 1 Details: 14 pmol; fMet-tRNA(fMet) at the P site and Phe-tRNA(phe) at the A site Classification: TRANSFER / Structure: DOUBLE HELIX / Synthetic?: No |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #2: EF-Tu
| Macromolecule | Name: EF-Tu / type: ligand / ID: 2 / Details: 20 pmol / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #3: GDP
| Macromolecule | Name: GDP / type: ligand / ID: 3 Details: 8 ul of 2.5mM; [Alpha-(32)P]GTP The radioactivity contributed by the Alpha-(32)P moiety of the GDP served to calculate a ~50% occupancy for the elongation factor. Recombinant expression: No |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #4: kirromycin
| Macromolecule | Name: kirromycin / type: ligand / ID: 4 / Details: 3 ul of 100 mM; antibiotic / Recombinant expression: No |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Chemical component information | 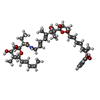 ChemComp-KIR: |
-Macromolecule #5: MF-mRNA
| Macromolecule | Name: MF-mRNA / type: ligand / ID: 5 / Details: 15 pmol; M(Met).F(Phe)-mRNA / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 20 mM HEPES-KOH, 6mM MgCl2, 150mM NH4Cl, 4mM 2-metcaptoethanol, 0.05 mM spermine and 2 mM spermidine |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 30 % / Chamber temperature: 93 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: two side blotting plunger Method: Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS EM420 |
|---|---|
| Electron beam | Acceleration voltage: 100 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 3.844 µm / Nominal defocus min: 1.041 µm / Nominal magnification: 49000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 3.844 µm / Nominal defocus min: 1.041 µm / Nominal magnification: 49000 |
| Sample stage | Specimen holder: Cryo transfer / Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Average: 93 K |
| Details | Microscope: Philips EM420 Imaging date: 1997 |
| Date | Jun 1, 1998 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 14 µm / Number real images: 45 / Average electron dose: 10 e/Å2 / Od range: 1.2 / Bits/pixel: 12 |
- Image processing
Image processing
| CTF correction | Details: defocus groups |
|---|---|
| Final two d classification | Number classes: 2 |
| Final angle assignment | Details: SPIDER definition |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 16.8 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER Details: Final map was calculated from 8 CTF corrected defocus groups Number images used: 7985 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: manual |
| Details | Protocol: Rigid Body. The EF-Tu domains were separately fitted by manual docking using program O (pdb codes for fitted coordinates: 1LS2, 1LU3) |
| Refinement | Protocol: RIGID BODY FIT / Target criteria: cross correlation coefficient |
| Output model |  PDB-1ls2:  PDB-1lu3: |
 Movie
Movie Controller
Controller



















