[English] 日本語
 Yorodumi
Yorodumi- EMDB-7774: 3.9A Cryo-EM structure of murine antibody bound at a novel epitop... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7774 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3.9A Cryo-EM structure of murine antibody bound at a novel epitope of respiratory syncytial virus fusion protein | |||||||||
 Map data Map data | 3.9A density map of R4.C6 in complex with RSV F710 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of syncytium formation by virus / host cell Golgi membrane / entry receptor-mediated virion attachment to host cell / symbiont entry into host cell / fusion of virus membrane with host plasma membrane /  viral envelope / host cell plasma membrane / virion membrane / viral envelope / host cell plasma membrane / virion membrane /  membrane / identical protein binding membrane / identical protein bindingSimilarity search - Function | |||||||||
| Biological species |   Mus musculus (house mouse) / Mus musculus (house mouse) /   Human respiratory syncytial virus A2 / Human respiratory syncytial virus A2 /    Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.9 Å cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Xie Q / Wang Z / Chen X / Ni F / Ma J / Wang Q | |||||||||
 Citation Citation |  Journal: PLoS One / Year: 2019 Journal: PLoS One / Year: 2019Title: Structure basis of neutralization by a novel site II/IV antibody against respiratory syncytial virus fusion protein. Authors: Qingqing Xie / Zhao Wang / Fengyun Ni / Xiaorui Chen / Jianpeng Ma / Nita Patel / Hanxin Lu / Ye Liu / Jing-Hui Tian / David Flyer / Michael J Massare / Larry Ellingsworth / Gregory Glenn / ...Authors: Qingqing Xie / Zhao Wang / Fengyun Ni / Xiaorui Chen / Jianpeng Ma / Nita Patel / Hanxin Lu / Ye Liu / Jing-Hui Tian / David Flyer / Michael J Massare / Larry Ellingsworth / Gregory Glenn / Gale Smith / Qinghua Wang /  Abstract: Globally, human respiratory syncytial virus (RSV) is a leading cause of lower respiratory tract infections in newborns, young children, and the elderly for which there is no vaccine. The RSV fusion ...Globally, human respiratory syncytial virus (RSV) is a leading cause of lower respiratory tract infections in newborns, young children, and the elderly for which there is no vaccine. The RSV fusion (F) glycoprotein is a major target for vaccine development. Here, we describe a novel monoclonal antibody (designated as R4.C6) that recognizes both pre-fusion and post-fusion RSV F, and binds with nanomole affinity to a unique neutralizing site comprised of antigenic sites II and IV on the globular head. A 3.9 Å-resolution structure of RSV F-R4.C6 Fab complex was obtained by single particle cryo-electron microscopy and 3D reconstruction. The structure unraveled detailed interactions of R4.C6 with antigenic site II on one protomer and site IV on a neighboring protomer of post-fusion RSV F protein. These findings significantly further our understanding of the antigenic complexity of the F protein and provide new insights into RSV vaccine design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7774.map.gz emd_7774.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7774-v30.xml emd-7774-v30.xml emd-7774.xml emd-7774.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
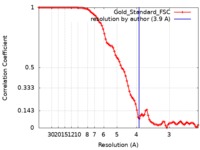
| FSC (resolution estimation) |  emd_7774_fsc.xml emd_7774_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_7774.png emd_7774.png | 47.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7774 http://ftp.pdbj.org/pub/emdb/structures/EMD-7774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7774 | HTTPS FTP |
-Related structure data
| Related structure data |  6cxcMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7774.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7774.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3.9A density map of R4.C6 in complex with RSV F710 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2546 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of R4.C6 Fab with RSV F protein
| Entire | Name: Complex of R4.C6 Fab with RSV F protein |
|---|---|
| Components |
|
-Supramolecule #1: Complex of R4.C6 Fab with RSV F protein
| Supramolecule | Name: Complex of R4.C6 Fab with RSV F protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: R4.C6 Fab
| Supramolecule | Name: R4.C6 Fab / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Recombinant expression | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Supramolecule #3: RSV F protein
| Supramolecule | Name: RSV F protein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:   Human respiratory syncytial virus A2 Human respiratory syncytial virus A2 |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
-Macromolecule #1: R4.C6 Fab Heavy Chain
| Macromolecule | Name: R4.C6 Fab Heavy Chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 12.831376 KDa |
| Recombinant expression | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Sequence | String: MAEVQLQQSG PELVKPGASV KISCKASGYA FSNSWMSWVK QRPGKGLEWI GRLFPADGDI TYNGHFKDKA ALTADKSSNT AYIQLSSLT SEDSAVYFCA RMDNSEVFWG QGTLVTVSA |
-Macromolecule #2: R4.C6 Fab Light Chain
| Macromolecule | Name: R4.C6 Fab Light Chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 13.457151 KDa |
| Recombinant expression | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Sequence | String: MADILLTQSQ KFMSTSVGDR VSITCKASQN VRTGVSWYQR KPGQSPKALI YLASNRHTGV PDRFTGRGSG TDFTLTISEV QSEDLADYF CLQHWTVPYT FGGGTKLEIK RTAAAPSRAN SFK |
-Macromolecule #3: Fusion glycoprotein F0, Envelope glycoprotein chimera
| Macromolecule | Name: Fusion glycoprotein F0, Envelope glycoprotein chimera / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 60.728047 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: QNITEEFYQS TCSAVSKGYL SALRTGWYTS VITIELSNIK ENKCNGTDAK VKLIKQELDK YKNAVTELQL LMQSTPATNN RARRELPRF MNYTLNNAKK TNVTLSKKQK QQFLGFLLGV GSAIASGVAV SKVLHLEGEV NKIKSALLST NKAVVSLSNG V SVLTSKVL ...String: QNITEEFYQS TCSAVSKGYL SALRTGWYTS VITIELSNIK ENKCNGTDAK VKLIKQELDK YKNAVTELQL LMQSTPATNN RARRELPRF MNYTLNNAKK TNVTLSKKQK QQFLGFLLGV GSAIASGVAV SKVLHLEGEV NKIKSALLST NKAVVSLSNG V SVLTSKVL DLKNYIDKQL LPIVNKQSCS ISNIETVIEF QQKNNRLLEI TREFSVNAGV TTPVSTYMLT NSELLSLIND MP ITNDQKK LMSNNVQIVR QQSYSIMSII KEEVLAYVVQ LPLYGVIDTP CWKLHTSPLC TTNTKEGSNI CLTRTDRGWY CDN AGSVSF FPQAETCKVQ SNRVFCDTMN SLTLPSEVNL CNVDIFNPKY DCKIMTSKTD VSSSVITSLG AIVSCYGKTK CTAS NKNRG IIKTFSNGCD YVSNKGVDTV SVGNTLYYVN KQEGKSLYVK GEPIINFYDP LVFPSDEFDA SISQVNEKIN QSLAF IRKS DELLHNVNAG KSTTNIMGAL VPRGSPGSGY IPEAPRDGQA YVRKDGEWVL LSTFLGSSHH HHHH |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: GRAPHENE OXIDE |
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 98 % / Chamber temperature: 295 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 4.7 mm Bright-field microscopy / Cs: 4.7 mm |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 10.0 sec. / Average electron dose: 50.0 e/Å2 |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6cxc: |
 Movie
Movie Controller
Controller