[English] 日本語
 Yorodumi
Yorodumi- EMDB-6332: 3D reconstruction of a ferritin-based nanoparticle displaying H1 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6332 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
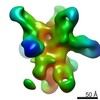
| Title | 3D reconstruction of a ferritin-based nanoparticle displaying H1 Hemagglutinin stem epitopes | |||||||||
 Map data Map data | ferritin-hemagglutinin nanoparticle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  nanoparticle / nanoparticle /  vaccine / epitope display / vaccine / epitope display /  influenza influenza | |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 16.0 Å cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Gallagher JR / Harris AK / Yassine HM / Boyington JC / McTamney PM / Wei CJ / Masaru M / Kong WP / Wang L / Zhang Y ...Gallagher JR / Harris AK / Yassine HM / Boyington JC / McTamney PM / Wei CJ / Masaru M / Kong WP / Wang L / Zhang Y / Joyce MG / Lingwood D / Moin SM / Andersen H / Okuno Y / Rao SS / Kwong PD / Mascola JR / Nabel GJ / Graham BS | |||||||||
 Citation Citation |  Journal: Nat Med / Year: 2015 Journal: Nat Med / Year: 2015Title: Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Authors: Hadi M Yassine / Jeffrey C Boyington / Patrick M McTamney / Chih-Jen Wei / Masaru Kanekiyo / Wing-Pui Kong / John R Gallagher / Lingshu Wang / Yi Zhang / M Gordon Joyce / Daniel Lingwood / ...Authors: Hadi M Yassine / Jeffrey C Boyington / Patrick M McTamney / Chih-Jen Wei / Masaru Kanekiyo / Wing-Pui Kong / John R Gallagher / Lingshu Wang / Yi Zhang / M Gordon Joyce / Daniel Lingwood / Syed M Moin / Hanne Andersen / Yoshinobu Okuno / Srinivas S Rao / Audray K Harris / Peter D Kwong / John R Mascola / Gary J Nabel / Barney S Graham /   Abstract: The antibody response to influenza is primarily focused on the head region of the hemagglutinin (HA) glycoprotein, which in turn undergoes antigenic drift, thus necessitating annual updates of ...The antibody response to influenza is primarily focused on the head region of the hemagglutinin (HA) glycoprotein, which in turn undergoes antigenic drift, thus necessitating annual updates of influenza vaccines. In contrast, the immunogenically subdominant stem region of HA is highly conserved and recognized by antibodies capable of binding multiple HA subtypes. Here we report the structure-based development of an H1 HA stem-only immunogen that confers heterosubtypic protection in mice and ferrets. Six iterative cycles of structure-based design (Gen1-Gen6) yielded successive H1 HA stabilized-stem (HA-SS) immunogens that lack the immunodominant head domain. Antigenic characterization, determination of two HA-SS crystal structures in complex with stem-specific monoclonal antibodies and cryo-electron microscopy analysis of HA-SS on ferritin nanoparticles (H1-SS-np) confirmed the preservation of key structural elements. Vaccination of mice and ferrets with H1-SS-np elicited broadly cross-reactive antibodies that completely protected mice and partially protected ferrets against lethal heterosubtypic H5N1 influenza virus challenge despite the absence of detectable H5N1 neutralizing activity in vitro. Passive transfer of immunoglobulin from H1-SS-np-immunized mice to naive mice conferred protection against H5N1 challenge, indicating that vaccine-elicited HA stem-specific antibodies can protect against diverse group 1 influenza strains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6332.map.gz emd_6332.map.gz | 7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6332-v30.xml emd-6332-v30.xml emd-6332.xml emd-6332.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6332_fsc.xml emd_6332_fsc.xml | 5.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_6332.png emd_6332.png | 124.6 KB | ||
| Masks |  emd_6332_msk_1.map emd_6332_msk_1.map | 8 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6332 http://ftp.pdbj.org/pub/emdb/structures/EMD-6332 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6332 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6332 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6332.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6332.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ferritin-hemagglutinin nanoparticle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.63 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Segmentation: relion postprocess automask: threshold=0.008 extend=4, soft-edge=8,
| Annotation | relion_postprocess automask: threshold=0.008 extend=4, soft-edge=8, | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| File |  emd_6332_msk_1.map emd_6332_msk_1.map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : H. pylori ferritin fusion with engineered H1 hemagglutin stem domain
| Entire | Name: H. pylori ferritin fusion with engineered H1 hemagglutin stem domain |
|---|---|
| Components |
|
-Supramolecule #1000: H. pylori ferritin fusion with engineered H1 hemagglutin stem domain
| Supramolecule | Name: H. pylori ferritin fusion with engineered H1 hemagglutin stem domain type: sample / ID: 1000 / Oligomeric state: nanoparticle of 24 subunits / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 1 MDa / Theoretical: 970 KDa / Method: gel filtration |
-Macromolecule #1: Hemagglutinin stem ferritin nanoparticle
| Macromolecule | Name: Hemagglutinin stem ferritin nanoparticle / type: protein_or_peptide / ID: 1 / Name.synonym: HA-stem ferritin nanoparticle / Number of copies: 24 / Oligomeric state: nanoparticle of 24 subunits / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Experimental: 1 MDa / Theoretical: 970 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: 293F Homo sapiens (human) / Recombinant cell: 293F |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 12 mM Na/K phosphate, 137 mM NaCl |
| Grid | Details: holey carbon film, Quantifoil |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 118 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 7.0 µm / Nominal defocus min: 3.5 µm / Nominal magnification: 75000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 7.0 µm / Nominal defocus min: 3.5 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Temperature | Min: 80 K / Max: 105 K / Average: 100 K |
| Date | Oct 1, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 209 / Average electron dose: 15 e/Å2 / Bits/pixel: 16 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X










