+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5429 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
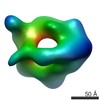
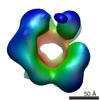
| Title | Negative stain 3D EM of MCM2-7 from Encephalitozoon cuniculi | |||||||||
 Map data Map data | Single-particle 3D reconstruction of EcuMCM2-7 with ATPgammaS | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  DNA replication / replicative helicase / DNA replication / replicative helicase /  MCM2-7 / MCM2-7 /  ATPase / hexameric motor ATPase / hexameric motor | |||||||||
| Biological species |   Encephalitozoon cuniculi (fungus) Encephalitozoon cuniculi (fungus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 24.0 Å negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Lyubimov AY / Costa A / Bleichert F / Botchan MR / Berger JM | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2012 Journal: Proc Natl Acad Sci U S A / Year: 2012Title: ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Authors: Artem Y Lyubimov / Alessandro Costa / Franziska Bleichert / Michael R Botchan / James M Berger /  Abstract: The heterohexameric minichromosome maintenance (MCM2-7) complex is an ATPase that serves as the central replicative helicase in eukaryotes. During initiation, the ring-shaped MCM2-7 particle is ...The heterohexameric minichromosome maintenance (MCM2-7) complex is an ATPase that serves as the central replicative helicase in eukaryotes. During initiation, the ring-shaped MCM2-7 particle is thought to open to facilitate loading onto DNA. The conformational state accessed during ring opening, the interplay between ATP binding and MCM2-7 architecture, and the use of these events in the regulation of DNA unwinding are poorly understood. To address these issues in isolation from the regulatory complexity of existing eukaryotic model systems, we investigated the structure/function relationships of a naturally minimized MCM2-7 complex from the microsporidian parasite Encephalitozoon cuniculi. Electron microscopy and small-angle X-ray scattering studies show that, in the absence of ATP, MCM2-7 spontaneously adopts a left-handed, open-ring structure. Nucleotide binding does not promote ring closure but does cause the particle to constrict in a two-step process that correlates with the filling of high- and low-affinity ATPase sites. Our findings support the idea that an open ring forms the default conformational state of the isolated MCM2-7 complex, and they provide a structural framework for understanding the multiphasic ATPase kinetics observed in different MCM2-7 systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5429.map.gz emd_5429.map.gz | 869 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5429-v30.xml emd-5429-v30.xml emd-5429.xml emd-5429.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5429.png emd_5429.png | 94.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5429 http://ftp.pdbj.org/pub/emdb/structures/EMD-5429 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5429 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5429 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5429.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5429.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle 3D reconstruction of EcuMCM2-7 with ATPgammaS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.56 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Encephalitozoon cuniculi MCM2-7 complex bound to ATPgammaS
| Entire | Name: Encephalitozoon cuniculi MCM2-7 complex bound to ATPgammaS |
|---|---|
| Components |
|
-Supramolecule #1000: Encephalitozoon cuniculi MCM2-7 complex bound to ATPgammaS
| Supramolecule | Name: Encephalitozoon cuniculi MCM2-7 complex bound to ATPgammaS type: sample / ID: 1000 / Oligomeric state: MCM2-7 heterohexamer / Number unique components: 6 |
|---|---|
| Molecular weight | Theoretical: 480 KDa |
-Macromolecule #1: Minichromosome maintenance protein 2
| Macromolecule | Name: Minichromosome maintenance protein 2 / type: protein_or_peptide / ID: 1 / Name.synonym: Mcm2 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Encephalitozoon cuniculi (fungus) Encephalitozoon cuniculi (fungus) |
| Molecular weight | Theoretical: 88 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac |
-Macromolecule #2: Minichromosome maintenance protein 3
| Macromolecule | Name: Minichromosome maintenance protein 3 / type: protein_or_peptide / ID: 2 / Name.synonym: Mcm3 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Encephalitozoon cuniculi (fungus) Encephalitozoon cuniculi (fungus) |
| Molecular weight | Theoretical: 77 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac |
-Macromolecule #3: Minichromosome maintenance protein 4
| Macromolecule | Name: Minichromosome maintenance protein 4 / type: protein_or_peptide / ID: 3 / Name.synonym: Mcm4 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Encephalitozoon cuniculi (fungus) Encephalitozoon cuniculi (fungus) |
| Molecular weight | Theoretical: 80 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac |
-Macromolecule #4: Minichromosome maintenance protein 5
| Macromolecule | Name: Minichromosome maintenance protein 5 / type: protein_or_peptide / ID: 4 / Name.synonym: Mcm5 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Encephalitozoon cuniculi (fungus) Encephalitozoon cuniculi (fungus) |
| Molecular weight | Theoretical: 79 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac |
-Macromolecule #5: Minichromosome maintenance protein 6
| Macromolecule | Name: Minichromosome maintenance protein 6 / type: protein_or_peptide / ID: 5 / Name.synonym: Mcm6 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Encephalitozoon cuniculi (fungus) Encephalitozoon cuniculi (fungus) |
| Molecular weight | Theoretical: 82 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac |
-Macromolecule #6: Minichromosome maintenance protein 7
| Macromolecule | Name: Minichromosome maintenance protein 7 / type: protein_or_peptide / ID: 6 / Name.synonym: Mcm7 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Encephalitozoon cuniculi (fungus) Encephalitozoon cuniculi (fungus) |
| Molecular weight | Theoretical: 77 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac Spodoptera frugiperda (fall armyworm) / Recombinant plasmid: pFastBac |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 300 mM NaOAc, 50 mM imidazole, pH 8.0, 5 mM Mg(OAc)2, 0.2 mM TCEP, 10% glycerol, 10 mM ATPgammaS |
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein (diluted to 0.03 mg/mL) floated on 2% uranyl formate for 10 sec (x 5 times) |
| Grid | Details: 400 mesh copper grids, nitrocellulose amyl acetate support, glow discharged, holey |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 30000 Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Date | Jan 5, 2011 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN / Number real images: 16532 / Average electron dose: 20 e/Å2 |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN2, SPARX Details: Nine iterative rounds were performed, with the output of each round used as starting model (after filtering) for the subsequent round until convergence. Convergence was determined by lack of ...Details: Nine iterative rounds were performed, with the output of each round used as starting model (after filtering) for the subsequent round until convergence. Convergence was determined by lack of further changes in volume features and distortion of FSC at higher resolution. Number images used: 16532 |
|---|---|
| Details | Particles selected automatically using DoG Picker and Batchboxer in the APPION environment |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid body. The 6 protomers were fit using sequential volume-to-volume fitting in Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller