[English] 日本語
 Yorodumi
Yorodumi- EMDB-42829: Straight actin filament from Arp2/3 branch junction sample (ADP) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Straight actin filament from Arp2/3 branch junction sample (ADP) | ||||||||||||
 Map data Map data | Helical F-actin from Arp2/3 branch complex (ADP) | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  actin / actin /  arp2/3 / arp2/3 /  cytoskeleton / branch / cytoskeleton / branch /  CYTOSOLIC PROTEIN CYTOSOLIC PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationcytoskeletal motor activator activity /  tropomyosin binding / tropomyosin binding /  myosin heavy chain binding / mesenchyme migration / myosin heavy chain binding / mesenchyme migration /  troponin I binding / actin filament bundle / filamentous actin / skeletal muscle thin filament assembly / actin filament bundle assembly / striated muscle thin filament ...cytoskeletal motor activator activity / troponin I binding / actin filament bundle / filamentous actin / skeletal muscle thin filament assembly / actin filament bundle assembly / striated muscle thin filament ...cytoskeletal motor activator activity /  tropomyosin binding / tropomyosin binding /  myosin heavy chain binding / mesenchyme migration / myosin heavy chain binding / mesenchyme migration /  troponin I binding / actin filament bundle / filamentous actin / skeletal muscle thin filament assembly / actin filament bundle assembly / striated muscle thin filament / skeletal muscle myofibril / actin monomer binding / skeletal muscle fiber development / troponin I binding / actin filament bundle / filamentous actin / skeletal muscle thin filament assembly / actin filament bundle assembly / striated muscle thin filament / skeletal muscle myofibril / actin monomer binding / skeletal muscle fiber development /  stress fiber / stress fiber /  titin binding / actin filament polymerization / titin binding / actin filament polymerization /  filopodium / filopodium /  actin filament / actin filament /  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding /  lamellipodium / lamellipodium /  cell body / cell body /  hydrolase activity / protein domain specific binding / hydrolase activity / protein domain specific binding /  calcium ion binding / positive regulation of gene expression / magnesium ion binding / calcium ion binding / positive regulation of gene expression / magnesium ion binding /  ATP binding / identical protein binding / ATP binding / identical protein binding /  cytoplasm cytoplasmSimilarity search - Function | ||||||||||||
| Biological species |   Gallus gallus (chicken) / Gallus gallus (chicken) /   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) | ||||||||||||
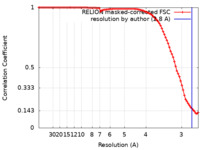
| Method | helical reconstruction /  cryo EM / Resolution: 2.8 Å cryo EM / Resolution: 2.8 Å | ||||||||||||
 Authors Authors | Chavali SS / Chou SZ / Sindelar CV | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex. Authors: Sai Shashank Chavali / Steven Z Chou / Wenxiang Cao / Thomas D Pollard / Enrique M De La Cruz / Charles V Sindelar /  Abstract: Arp2/3 complex nucleates branched actin filaments for cell and organelle movements. Here we report a 2.7 Å resolution cryo-EM structure of the mature branch junction formed by S. pombe Arp2/3 ...Arp2/3 complex nucleates branched actin filaments for cell and organelle movements. Here we report a 2.7 Å resolution cryo-EM structure of the mature branch junction formed by S. pombe Arp2/3 complex that provides details about interactions with both mother and daughter filaments. We determine a second structure at 3.2 Å resolution with the phosphate analog BeF bound with ADP to Arp3 and ATP bound to Arp2. In this ADP-BeF transition state the outer domain of Arp3 is rotated 2° toward the mother filament compared with the ADP state and makes slightly broader contacts with actin in both the mother and daughter filaments. Thus, dissociation of P from the ADP-P transition state reduces the interactions of Arp2/3 complex with the actin filaments and may contribute to the lower mechanical stability of mature branch junctions with ADP bound to the Arps. Our structures also reveal that the mother filament in contact with Arp2/3 complex is slightly bent and twisted, consistent with the preference of Arp2/3 complex binding curved actin filaments. The small degree of twisting constrains models of actin filament mechanics. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42829.map.gz emd_42829.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42829-v30.xml emd-42829-v30.xml emd-42829.xml emd-42829.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42829_fsc.xml emd_42829_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_42829.png emd_42829.png | 77.4 KB | ||
| Filedesc metadata |  emd-42829.cif.gz emd-42829.cif.gz | 6.7 KB | ||
| Others |  emd_42829_half_map_1.map.gz emd_42829_half_map_1.map.gz emd_42829_half_map_2.map.gz emd_42829_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42829 http://ftp.pdbj.org/pub/emdb/structures/EMD-42829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42829 | HTTPS FTP |
-Related structure data
| Related structure data |  8uz0MC  8uxwC  8uxxC  8uz1C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42829.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42829.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical F-actin from Arp2/3 branch complex (ADP) | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.346 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Helical F-actin from Arp2/3 branch complex (ADP)
| File | emd_42829_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical F-actin from Arp2/3 branch complex (ADP) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Helical F-actin from Arp2/3 branch complex (ADP)
| File | emd_42829_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical F-actin from Arp2/3 branch complex (ADP) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Actin
| Entire | Name: Actin |
|---|---|
| Components |
|
-Supramolecule #1: Actin
| Supramolecule | Name: Actin / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Actin filament |
|---|---|
| Source (natural) | Organism:   Gallus gallus (chicken) Gallus gallus (chicken) |
-Macromolecule #1: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 1 / Number of copies: 9 / Enantiomer: LEVO EC number:  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) |
| Molecular weight | Theoretical: 41.51634 KDa |
| Sequence | String: ETTALVCDNG SGLVKAGFAG DDAPRAVFPS IVGRPRHQGV MVGMGQKDSY VGDEAQSKRG ILTLKYPIE(HIC) GIITNW DDM EKIWHHTFYN ELRVAPEEHP TLLTEAPLNP KANREKMTQI MFETFNVPAM YVAIQAVLSL YASGRTTGIV LDSGDGV TH NVPIYEGYAL ...String: ETTALVCDNG SGLVKAGFAG DDAPRAVFPS IVGRPRHQGV MVGMGQKDSY VGDEAQSKRG ILTLKYPIE(HIC) GIITNW DDM EKIWHHTFYN ELRVAPEEHP TLLTEAPLNP KANREKMTQI MFETFNVPAM YVAIQAVLSL YASGRTTGIV LDSGDGV TH NVPIYEGYAL PHAIMRLDLA GRDLTDYLMK ILTERGYSFV TTAEREIVRD IKEKLCYVAL DFENEMATAA SSSSLEKS Y ELPDGQVITI GNERFRCPET LFQPSFIGME SAGIHETTYN SIMKCDIDIR KDLYANNVMS GGTTMYPGIA DRMQKEITA LAPSTMKIKI IAPPERKYSV WIGGSILASL STFQQMWITK QEYDEAGPSI VHRKCF UniProtKB:  Actin, alpha skeletal muscle Actin, alpha skeletal muscle |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 9 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 9 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: The samples were incubated on the grid for 50 s and the extra solution was blotted using two Vitrobot filter papers (0.55/20 mm, Grade 595, Ted Pella) for 4 s at 0 blot force. The grids were ...Details: The samples were incubated on the grid for 50 s and the extra solution was blotted using two Vitrobot filter papers (0.55/20 mm, Grade 595, Ted Pella) for 4 s at 0 blot force. The grids were plunged into liquid ethane at ~180 degrees C with a wait time of 0.5 s.. |
| Details | Actin monomers with bound Ca2+ were converted to Mg2+-actin by equilibrating with 50 micromolar MgCl2 and 0.2 mM EGTA (pH 7.5) for 10 min on ice. Actin was polymerized in the presence of capping protein (CP) by sequentially mixing 8.75 micromolar Mg-ATP-actin monomers with 0.75 micromolar CP and equilibrated at room temperature for 1 h. In parallel, Arp2/3 complex (0.4 micromolar) in QB buffer was activated by mixing 0.85 micromolar GCN4-VCA and 50 micromolar ATP and incubated at 4 degrees for 1 h. The capped actin filaments sample was then gently mixed with an equal volume of activated Arp2/3 complex sample using cut pipette tips and equilibrated at 4 degrees for 5 min. Daughter filaments were formed and elongated in the presence of CP by adding 0.25 micromolar Mg-actin monomers, 50 micromolar ATP, and 40 nM CP and incubated for 5 min at room temperature. This step was subsequently repeated 4 more times, then aged for ~90 min before preparing grids for cryo-EM data collection. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 64000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 64000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV Details: Electron micrographs for image reconstructions were collected using Titan Krios equipped with X-cold field emission gun at 300 kV, Gatan image filter with slit width of 20 eV and a nanoprobe. |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Details | The vitrified grids were screened for sample homogeneity and ice thickness in a Glacios 200 kV transmission electron microscope equipped with Gatan K2 summit camera. Electron micrographs for image reconstructions were collected using Titan Krios equipped with X-cold field emission gun at 300 kV, Gatan image filter with slit width of 20 eV and a nanoprobe. |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 8519 / Average exposure time: 3.3 sec. / Average electron dose: 51.4 e/Å2 Details: Each movie contains 41 frames with a frame time of 0.08 s. A dose rate of 28.4 counts/pixel/s and a physical pixel size of 1.346 Angstroms was used. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X