[English] 日本語
 Yorodumi
Yorodumi- EMDB-4230: Single particle cryo em structure of Mycobacterium tuberculosis R... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4230 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle cryo em structure of Mycobacterium tuberculosis RNA polymerase in complex with Fidaxomicin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lipiarmycin / RNA pol /  RNAP / RNAP /  inhibitor / inhibitor /  drug / Clostridium difficile / drug / Clostridium difficile /  ANTIBIOTIC / Tiacumicin B / CCDC 114782 / ANTIBIOTIC / Tiacumicin B / CCDC 114782 /  transcription transcription | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to water / Antimicrobial action and antimicrobial resistance in Mtb /  sigma factor activity / peptidoglycan-based cell wall / sigma factor activity / peptidoglycan-based cell wall /  DNA-directed RNA polymerase complex / DNA-templated transcription initiation / DNA-directed RNA polymerase complex / DNA-templated transcription initiation /  ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity / ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity /  DNA-directed RNA polymerase / DNA-directed RNA polymerase /  protein dimerization activity ...response to water / Antimicrobial action and antimicrobial resistance in Mtb / protein dimerization activity ...response to water / Antimicrobial action and antimicrobial resistance in Mtb /  sigma factor activity / peptidoglycan-based cell wall / sigma factor activity / peptidoglycan-based cell wall /  DNA-directed RNA polymerase complex / DNA-templated transcription initiation / DNA-directed RNA polymerase complex / DNA-templated transcription initiation /  ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity / ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity /  DNA-directed RNA polymerase / DNA-directed RNA polymerase /  protein dimerization activity / response to antibiotic / DNA-templated transcription / magnesium ion binding / protein dimerization activity / response to antibiotic / DNA-templated transcription / magnesium ion binding /  DNA binding / zinc ion binding / DNA binding / zinc ion binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) | |||||||||
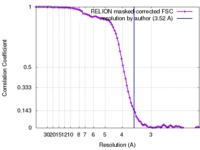
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.52 Å cryo EM / Resolution: 3.52 Å | |||||||||
 Authors Authors | Das K | |||||||||
| Funding support |  Belgium, 1 items Belgium, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2018 Journal: Mol Cell / Year: 2018Title: Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3). Authors: Wei Lin / Kalyan Das / David Degen / Abhishek Mazumder / Diego Duchi / Dongye Wang / Yon W Ebright / Richard Y Ebright / Elena Sineva / Matthew Gigliotti / Aashish Srivastava / Sukhendu ...Authors: Wei Lin / Kalyan Das / David Degen / Abhishek Mazumder / Diego Duchi / Dongye Wang / Yon W Ebright / Richard Y Ebright / Elena Sineva / Matthew Gigliotti / Aashish Srivastava / Sukhendu Mandal / Yi Jiang / Yu Liu / Ruiheng Yin / Zhening Zhang / Edward T Eng / Dennis Thomas / Stefano Donadio / Haibo Zhang / Changsheng Zhang / Achillefs N Kapanidis / Richard H Ebright /      Abstract: Fidaxomicin is an antibacterial drug in clinical use for treatment of Clostridium difficile diarrhea. The active ingredient of fidaxomicin, lipiarmycin A3 (Lpm), functions by inhibiting bacterial ...Fidaxomicin is an antibacterial drug in clinical use for treatment of Clostridium difficile diarrhea. The active ingredient of fidaxomicin, lipiarmycin A3 (Lpm), functions by inhibiting bacterial RNA polymerase (RNAP). Here we report a cryo-EM structure of Mycobacterium tuberculosis RNAP holoenzyme in complex with Lpm at 3.5-Å resolution. The structure shows that Lpm binds at the base of the RNAP "clamp." The structure exhibits an open conformation of the RNAP clamp, suggesting that Lpm traps an open-clamp state. Single-molecule fluorescence resonance energy transfer experiments confirm that Lpm traps an open-clamp state and define effects of Lpm on clamp dynamics. We suggest that Lpm inhibits transcription by trapping an open-clamp state, preventing simultaneous interaction with promoter -10 and -35 elements. The results account for the absence of cross-resistance between Lpm and other RNAP inhibitors, account for structure-activity relationships of Lpm derivatives, and enable structure-based design of improved Lpm derivatives. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4230.map.gz emd_4230.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4230-v30.xml emd-4230-v30.xml emd-4230.xml emd-4230.xml | 27.3 KB 27.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4230_fsc.xml emd_4230_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_4230.png emd_4230.png | 123.5 KB | ||
| Filedesc metadata |  emd-4230.cif.gz emd-4230.cif.gz | 8.9 KB | ||
| Others |  emd_4230_half_map_1.map.gz emd_4230_half_map_1.map.gz emd_4230_half_map_2.map.gz emd_4230_half_map_2.map.gz | 40.8 MB 40.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4230 http://ftp.pdbj.org/pub/emdb/structures/EMD-4230 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4230 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4230 | HTTPS FTP |
-Related structure data
| Related structure data |  6fbvMC  6asgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4230.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4230.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_4230_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
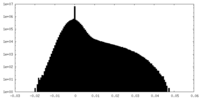
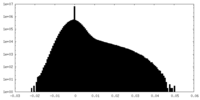
| Density Histograms |
-Half map: #2
| File | emd_4230_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
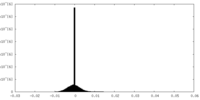
| Density Histograms |
- Sample components
Sample components
+Entire : structure of Mycobacterium tuberculosis RNA polymerase in complex...
+Supramolecule #1: structure of Mycobacterium tuberculosis RNA polymerase in complex...
+Macromolecule #1: DNA-directed RNA polymerase subunit alpha
+Macromolecule #2: DNA-directed RNA polymerase subunit beta
+Macromolecule #3: DNA-directed RNA polymerase subunit beta'
+Macromolecule #4: DNA-directed RNA polymerase subunit omega
+Macromolecule #5: RNA polymerase sigma factor SigA
+Macromolecule #6: ZINC ION
+Macromolecule #7: MAGNESIUM ION
+Macromolecule #8: Fidaxomicin
+Macromolecule #9: water
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Details: 3.5 microliter 1 microM Mtb RNAP-Lpm and 50 microMolar Lpm in 20 mM Tris-HCl, pH 8.0, 75 mM NaCl, 5 mM MgCl2, 5 mM dithiothreitol, and 0.1% n-octyl-beta-D-glucopyranoside |
|---|---|
| Grid | Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: LACEY / Support film - Film thickness: 100 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 291 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: -0.002 µm / Nominal defocus min: -0.001 µm / Nominal magnification: 130000 Bright-field microscopy / Nominal defocus max: -0.002 µm / Nominal defocus min: -0.001 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-35 / Number grids imaged: 2 / Number real images: 2458 / Average exposure time: 0.2 sec. / Average electron dose: 1.4 e/Å2 Details: Movies were recorded at 200 ms/frame for 10s (50 frames total), resulting in a total radiation dose of 72.05 electrons/A**2 per movie Defocus range was varied between 1.0 - 2.0 micrometer. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: OTHER / Overall B value: 95.3 / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-6fbv: |
 Movie
Movie Controller
Controller














 Z
Z Y
Y X
X