+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4174 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Ageratum Yellow Vein virus (AYVV) | ||||||||||||||||||
 Map data Map data | Ageratum Yellow Vein virus cryoEM density map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | AYVV / geminivirus /  ssDNA / gemini / ssDNA / gemini /  VIRUS VIRUS | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationT=1 icosahedral viral capsid / viral penetration into host nucleus / symbiont entry into host cell / host cell nucleus / structural molecule activity /  DNA binding / DNA binding /  metal ion binding metal ion bindingSimilarity search - Function | ||||||||||||||||||
| Biological species |   Ageratum yellow vein virus Ageratum yellow vein virus | ||||||||||||||||||
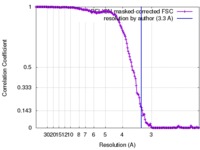
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.3 Å cryo EM / Resolution: 3.3 Å | ||||||||||||||||||
 Authors Authors | Hesketh EL / Saunders K | ||||||||||||||||||
| Funding support |  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: The 3.3 Å structure of a plant geminivirus using cryo-EM. Authors: Emma L Hesketh / Keith Saunders / Chloe Fisher / Joran Potze / John Stanley / George P Lomonossoff / Neil A Ranson /  Abstract: Geminiviruses are major plant pathogens that threaten food security globally. They have a unique architecture built from two incomplete icosahedral particles, fused to form a geminate capsid. ...Geminiviruses are major plant pathogens that threaten food security globally. They have a unique architecture built from two incomplete icosahedral particles, fused to form a geminate capsid. However, despite their importance to agricultural economies and fundamental biological interest, the details of how this is realized in 3D remain unknown. Here we report the structure of Ageratum yellow vein virus at 3.3 Å resolution, using single-particle cryo-electron microscopy, together with an atomic model that shows that the N-terminus of the single capsid protein (CP) adopts three different conformations essential for building the interface between geminate halves. Our map also contains density for ~7 bases of single-stranded DNA bound to each CP, and we show that the interactions between the genome and CPs are different at the interface than in the rest of the capsid. With additional mutagenesis data, this suggests a central role for DNA binding-induced conformational change in directing the assembly of geminate capsids. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4174.map.gz emd_4174.map.gz | 197.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4174-v30.xml emd-4174-v30.xml emd-4174.xml emd-4174.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4174_fsc.xml emd_4174_fsc.xml | 15.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4174.png emd_4174.png | 194.4 KB | ||
| Filedesc metadata |  emd-4174.cif.gz emd-4174.cif.gz | 6 KB | ||
| Others |  emd_4174_additional.map.gz emd_4174_additional.map.gz | 273.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4174 http://ftp.pdbj.org/pub/emdb/structures/EMD-4174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4174 | HTTPS FTP |
-Related structure data
| Related structure data |  6f2sMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4174.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4174.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ageratum Yellow Vein virus cryoEM density map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Ageratum Yellow Vein virus unsharpened
| File | emd_4174_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ageratum Yellow Vein virus unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ageratum yellow vein virus
| Entire | Name:   Ageratum yellow vein virus Ageratum yellow vein virus |
|---|---|
| Components |
|
-Supramolecule #1: Ageratum yellow vein virus
| Supramolecule | Name: Ageratum yellow vein virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 / NCBI-ID: 44560 / Sci species name: Ageratum yellow vein virus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:   Ageratum (plant) Ageratum (plant) |
| Molecular weight | Theoretical: 251 KDa |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Ageratum yellow vein virus Ageratum yellow vein virus |
| Molecular weight | Theoretical: 22.500703 KDa |
| Recombinant expression | Organism:   Nicotiana benthamiana (plant) Nicotiana benthamiana (plant) |
| Sequence | String: PDVPKGCEGP CKVQSYEQRH DISHVGKVLC VSDVTRGNGL THRVGKRFCV KSVYVLGKIW MDENIKTKNH TNTVMFYLVR DRRPFGTAM DFGQVFNMYD NEPSTATIKN DLRDRYQVLR KFTSTVTGGQ YASKEQALVK KFMKINNYVV YNHQEAAKYD N HTENALLL ...String: PDVPKGCEGP CKVQSYEQRH DISHVGKVLC VSDVTRGNGL THRVGKRFCV KSVYVLGKIW MDENIKTKNH TNTVMFYLVR DRRPFGTAM DFGQVFNMYD NEPSTATIKN DLRDRYQVLR KFTSTVTGGQ YASKEQALVK KFMKINNYVV YNHQEAAKYD N HTENALLL YMACTHASNP VYATLKIRIY FYDSVQN UniProtKB:  Capsid protein Capsid protein |
-Macromolecule #3: coat protein subunit I
| Macromolecule | Name: coat protein subunit I / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Ageratum yellow vein virus Ageratum yellow vein virus |
| Molecular weight | Theoretical: 23.644094 KDa |
| Recombinant expression | Organism:   Nicotiana benthamiana (plant) Nicotiana benthamiana (plant) |
| Sequence | String: RLYRMYRTPD VPKGCEGPCK VQSYEQRHDI SHVGKVLCVS DVTRGNGLTH RVGKRFCVKS VYVLGKIWMD ENIKTKNHTN TVMFYLVRD RRPFGTAMDF GQVFNMYDNE PSTATIKNDL RDRYQVLRKF TSTVTGGQYA SKEQALVKKF MKINNYVVYN H QEAAKYDN ...String: RLYRMYRTPD VPKGCEGPCK VQSYEQRHDI SHVGKVLCVS DVTRGNGLTH RVGKRFCVKS VYVLGKIWMD ENIKTKNHTN TVMFYLVRD RRPFGTAMDF GQVFNMYDNE PSTATIKNDL RDRYQVLRKF TSTVTGGQYA SKEQALVKKF MKINNYVVYN H QEAAKYDN HTENALLLYM ACTHASNPVY ATLKIRIYFY DSVQN UniProtKB:  Capsid protein Capsid protein |
-Macromolecule #4: coat protein subunit H
| Macromolecule | Name: coat protein subunit H / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Ageratum yellow vein virus Ageratum yellow vein virus |
| Molecular weight | Theoretical: 25.664473 KDa |
| Recombinant expression | Organism:   Nicotiana benthamiana (plant) Nicotiana benthamiana (plant) |
| Sequence | String: NRRRTWTNRP MYRKPRLYRM YRTPDVPKGC EGPCKVQSYE QRHDISHVGK VLCVSDVTRG NGLTHRVGKR FCVKSVYVLG KIWMDENIK TKNHTNTVMF YLVRDRRPFG TAMDFGQVFN MYDNEPSTAT IKNDLRDRYQ VLRKFTSTVT GGQYASKEQA L VKKFMKIN ...String: NRRRTWTNRP MYRKPRLYRM YRTPDVPKGC EGPCKVQSYE QRHDISHVGK VLCVSDVTRG NGLTHRVGKR FCVKSVYVLG KIWMDENIK TKNHTNTVMF YLVRDRRPFG TAMDFGQVFN MYDNEPSTAT IKNDLRDRYQ VLRKFTSTVT GGQYASKEQA L VKKFMKIN NYVVYNHQEA AKYDNHTENA LLLYMACTHA SNPVYATLKI RIYFYDSVQN UniProtKB:  Capsid protein Capsid protein |
-Macromolecule #2: ssDNA loop
| Macromolecule | Name: ssDNA loop / type: dna / ID: 2 / Number of copies: 10 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Ageratum yellow vein virus Ageratum yellow vein virus |
| Molecular weight | Theoretical: 2.05139 KDa |
| Sequence | String: (DC)(DA)(DA)(DC)(DC)(DA)(DC) |
-Macromolecule #5: ssDNA loop associated with subunit H
| Macromolecule | Name: ssDNA loop associated with subunit H / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Ageratum yellow vein virus Ageratum yellow vein virus |
| Molecular weight | Theoretical: 1.762208 KDa |
| Sequence | String: (DC)(DA)(DA)(DC)(DC)(DA) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Component - Concentration: 100.0 mM / Component - Formula: NaPo4 / Component - Name: Sodium phosphate |
|---|---|
| Grid | Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Support film - Film thickness: 2000 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.037 kPa / Details: PELCO easiglow |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 5.0 µm / Calibrated defocus min: 0.3 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 75000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 78.0 K / Max: 78.0 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 12028 / Average exposure time: 2.0 sec. / Average electron dose: 110.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6f2s: |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X