[English] 日本語
 Yorodumi
Yorodumi- EMDB-41062: CryoEM structure of an inward-facing MelBSt at a Na(+)-bound and ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of an inward-facing MelBSt at a Na(+)-bound and sugar low-affinity conformation | |||||||||
 Map data Map data | The model was refined against this map. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Sugar transporter / Cation-coupled symporter / Sugar transporter / Cation-coupled symporter /  Na(+) binding / Protein conformation / Na(+) binding / Protein conformation /  Nanobodies / NabFab / Nanobodies / NabFab /  CryoEM / CryoEM /  Membrane proteins / Membrane proteins /  protein-protein interaction / protein-protein interaction /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationorganic substance transport /  symporter activity / sodium ion transport / carbohydrate transport / symporter activity / sodium ion transport / carbohydrate transport /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli (E. coli) / Escherichia coli (E. coli) /   Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) / synthetic construct (others) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) / synthetic construct (others) | |||||||||
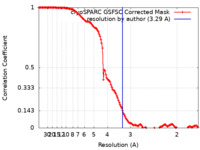
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.29 Å cryo EM / Resolution: 3.29 Å | |||||||||
 Authors Authors | Guan L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Mobile barrier mechanisms for Na-coupled symport in an MFS sugar transporter. Authors: Parameswaran Hariharan / Yuqi Shi / Satoshi Katsube / Katleen Willibal / Nathan D Burrows / Patrick Mitchell / Amirhossein Bakhtiiari / Samantha Stanfield / Els Pardon / H Ronald Kaback / ...Authors: Parameswaran Hariharan / Yuqi Shi / Satoshi Katsube / Katleen Willibal / Nathan D Burrows / Patrick Mitchell / Amirhossein Bakhtiiari / Samantha Stanfield / Els Pardon / H Ronald Kaback / Ruibin Liang / Jan Steyaert / Rosa Viner / Lan Guan /   Abstract: While many 3D structures of cation-coupled transporters have been determined, the mechanistic details governing the obligatory coupling and functional regulations still remain elusive. The bacterial ...While many 3D structures of cation-coupled transporters have been determined, the mechanistic details governing the obligatory coupling and functional regulations still remain elusive. The bacterial melibiose transporter (MelB) is a prototype of major facilitator superfamily transporters. With a conformation-selective nanobody, we determined a low-sugar affinity inward-facing Na-bound cryoEM structure. The available outward-facing sugar-bound structures showed that the N- and C-terminal residues of the inner barrier contribute to the sugar selectivity. The inward-open conformation shows that the sugar selectivity pocket is also broken when the inner barrier is broken. Isothermal titration calorimetry measurements revealed that this inward-facing conformation trapped by this nanobody exhibited a greatly decreased sugar-binding affinity, suggesting the mechanisms for substrate intracellular release and accumulation. While the inner/outer barrier shift directly regulates the sugar-binding affinity, it has little or no effect on the cation binding, which is supported by molecular dynamics simulations. Furthermore, the hydron/deuterium exchange mass spectrometry analyses allowed us to identify dynamic regions; some regions are involved in the functionally important inner barrier-specific salt-bridge network, which indicates their critical roles in the barrier switching mechanisms for transport. These complementary results provided structural and dynamic insights into the mobile barrier mechanism for cation-coupled symport. #1:  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Mobile barrier mechanisms for Na+-coupled symport in an MFS sugar transporter Authors: Hariharan P / Shi Y / Katsube S / Willibal K / Burrows ND / Mitchell P / Bakhtiiari A / Stanfield S / Pardon E / Kaback HR / Liang R / Steyaert J / Viner R / Gran L | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41062.map.gz emd_41062.map.gz | 198.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41062-v30.xml emd-41062-v30.xml emd-41062.xml emd-41062.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41062_fsc.xml emd_41062_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_41062.png emd_41062.png | 92.3 KB | ||
| Masks |  emd_41062_msk_1.map emd_41062_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41062.cif.gz emd-41062.cif.gz | 7.4 KB | ||
| Others |  emd_41062_additional_1.map.gz emd_41062_additional_1.map.gz emd_41062_half_map_1.map.gz emd_41062_half_map_1.map.gz emd_41062_half_map_2.map.gz emd_41062_half_map_2.map.gz | 107.5 MB 200.5 MB 200.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41062 http://ftp.pdbj.org/pub/emdb/structures/EMD-41062 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41062 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41062 | HTTPS FTP |
-Related structure data
| Related structure data |  8t60MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41062.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41062.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The model was refined against this map. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41062_msk_1.map emd_41062_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_41062_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_41062_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_41062_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MelBSt bound with Nb725_4-NabFab complex
| Entire | Name: MelBSt bound with Nb725_4-NabFab complex |
|---|---|
| Components |
|
-Supramolecule #1: MelBSt bound with Nb725_4-NabFab complex
| Supramolecule | Name: MelBSt bound with Nb725_4-NabFab complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: Melibiose permease
| Macromolecule | Name: Melibiose permease / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria)Strain: LT2 |
| Molecular weight | Theoretical: 54.104438 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: SISMTTKLSY GFGAFGKDFA IGIVYMYLMY YYTDVVGLSV GLVGTLFLVA RIWDAINDPI MGWIVNATRS RWGKFKPWIL IGTLTNSLV LFLLFSAHLF EGTAQVVFVC VTYILWGMTY TIMDIPFWSL VPTITLDKRE REQLVPFPRF FASLAGFVTA G ITLPFVSY ...String: SISMTTKLSY GFGAFGKDFA IGIVYMYLMY YYTDVVGLSV GLVGTLFLVA RIWDAINDPI MGWIVNATRS RWGKFKPWIL IGTLTNSLV LFLLFSAHLF EGTAQVVFVC VTYILWGMTY TIMDIPFWSL VPTITLDKRE REQLVPFPRF FASLAGFVTA G ITLPFVSY VGGADRGFGF QMFTLVLIAF FIASTIVTLR NVHEVYSSDN GVTAGRPHLT LKTIVGLIYK NDQLSCLLGM AL AYNIASN IINGFAIYYF TYVIGDADLF PYYLSYAGAA NLLTLIVFPR LVKMLSRRIL WAGASVMPVL SCAGLFAMAL ADI HNAALI VAAGIFLNIG TALFWVLQVI MVADTVDYGE FKLNIRCESI AYSVQTMVVK GGSAFAAFFI ALVLGLIGYT PNVA QSAQT LQGMQFIMIV LPVLFFMMTL VLYFRYYRLN GDMLRKIQIH LLDKYRKTPP FVEQPDSPAI SVVATSDVKA HHHHH HHHH H UniProtKB: Melibiose permease |
-Macromolecule #2: Nb725_4
| Macromolecule | Name: Nb725_4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 14.817506 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSQRQLVESG GGLVQPGGSL RLSCAVSGII FRDNAMGWYR QAPGKEREWV ATITDLGYTA YADSVKGRFT ISRDNAKDTV YLQMNSLEP EDTAVYYCHL PGTAAGDYWG KGTPVTVSSL EVLFQGPHHH HHHHH |
-Macromolecule #3: NabFab_H Chain
| Macromolecule | Name: NabFab_H Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 25.684463 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFSYYSIHWV RQAPGKGLEW VAYISSSSSY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARGYQYWQYH ASWYWNGGLD YWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFSYYSIHWV RQAPGKGLEW VAYISSSSSY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARGYQYWQYH ASWYWNGGLD YWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV TVSWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH T |
-Macromolecule #4: NabFab_L Chain
| Macromolecule | Name: NabFab_L Chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.258783 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSSSLITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSSSLITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #5: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 5 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI CETA (4k x 4k) / Number real images: 20778 / Average exposure time: 0.0535 sec. / Average electron dose: 1.28 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Protocol: AB INITIO MODEL | ||||||||||
| Output model |  PDB-8t60: |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X