+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Karyopherin-beta2 bound to HNRNPH2 PY-NLS | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | RNA-binding protein TNPO1 Cryo-EM KapB2 HNRNP /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationTristetraprolin (TTP, ZFP36) binds and destabilizes mRNA /  Intraflagellar transport / Postmitotic nuclear pore complex (NPC) reformation / nuclear import signal receptor activity / Intraflagellar transport / Postmitotic nuclear pore complex (NPC) reformation / nuclear import signal receptor activity /  nuclear localization sequence binding / nuclear localization sequence binding /  regulation of RNA splicing / Processing of Capped Intron-Containing Pre-mRNA / mRNA Splicing - Major Pathway / regulation of RNA splicing / Processing of Capped Intron-Containing Pre-mRNA / mRNA Splicing - Major Pathway /  cilium / cilium /  small GTPase binding ...Tristetraprolin (TTP, ZFP36) binds and destabilizes mRNA / small GTPase binding ...Tristetraprolin (TTP, ZFP36) binds and destabilizes mRNA /  Intraflagellar transport / Postmitotic nuclear pore complex (NPC) reformation / nuclear import signal receptor activity / Intraflagellar transport / Postmitotic nuclear pore complex (NPC) reformation / nuclear import signal receptor activity /  nuclear localization sequence binding / nuclear localization sequence binding /  regulation of RNA splicing / Processing of Capped Intron-Containing Pre-mRNA / mRNA Splicing - Major Pathway / regulation of RNA splicing / Processing of Capped Intron-Containing Pre-mRNA / mRNA Splicing - Major Pathway /  cilium / cilium /  small GTPase binding / protein import into nucleus / small GTPase binding / protein import into nucleus /  postsynaptic density / postsynaptic density /  ribonucleoprotein complex / ribonucleoprotein complex /  RNA binding / extracellular exosome / RNA binding / extracellular exosome /  nucleoplasm / nucleoplasm /  membrane / membrane /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.17 Å cryo EM / Resolution: 3.17 Å | |||||||||||||||
 Authors Authors | Gonzalez A / Fung HYJ / Chook YM | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: A new Karyopherin-β2 binding PY-NLS epitope of HNRNPH2 linked to neurodevelopmental disorders. Authors: Abner Gonzalez / Hong Joo Kim / Brian D Freibaum / Ho Yee Joyce Fung / Chad A Brautigam / J Paul Taylor / Yuh Min Chook /  Abstract: The HNRNPH2 proline-tyrosine nuclear localization signal (PY-NLS) is mutated in HNRNPH2-related X-linked neurodevelopmental disorder, causing the normally nuclear HNRNPH2 to accumulate in the ...The HNRNPH2 proline-tyrosine nuclear localization signal (PY-NLS) is mutated in HNRNPH2-related X-linked neurodevelopmental disorder, causing the normally nuclear HNRNPH2 to accumulate in the cytoplasm. We solved the cryoelectron microscopy (cryo-EM) structure of Karyopherin-β2/Transportin-1 bound to the HNRNPH2 PY-NLS to understand importin-NLS recognition and disruption in disease. HNRNPH2 RPGPY is a typical R-X-P-Y motif comprising PY-NLS epitopes 2 and 3, followed by an additional Karyopherin-β2-binding epitope, we term epitope 4, at residues DRP; no density is present for PY-NLS epitope 1. Disease variant mutations at epitopes 2-4 impair Karyopherin-β2 binding and cause aberrant cytoplasmic accumulation in cells, emphasizing the role of nuclear import defect in disease. Sequence/structure analysis suggests that strong PY-NLS epitopes 4 are rare and thus far limited to close paralogs of HNRNPH2, HNRNPH1, and HNRNPF. Epitope 4-binidng hotspot Karyopherin-β2 W373 corresponds to close paralog Karyopherin-β2b/Transportin-2 W370, a pathological variant site in neurodevelopmental abnormalities, suggesting that Karyopherin-β2b/Transportin-2-HNRNPH2/H1/F interactions may be compromised in the abnormalities. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40455.map.gz emd_40455.map.gz | 403.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40455-v30.xml emd-40455-v30.xml emd-40455.xml emd-40455.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40455.png emd_40455.png | 53 KB | ||
| Others |  emd_40455_half_map_1.map.gz emd_40455_half_map_1.map.gz emd_40455_half_map_2.map.gz emd_40455_half_map_2.map.gz | 764 MB 764 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40455 http://ftp.pdbj.org/pub/emdb/structures/EMD-40455 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40455 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40455 | HTTPS FTP |
-Related structure data
| Related structure data |  8sghMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40455.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40455.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.415 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_40455_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
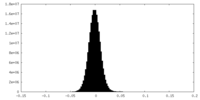
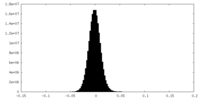
| Density Histograms |
-Half map: #1
| File | emd_40455_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
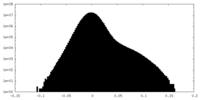
| Density Histograms |
- Sample components
Sample components
-Entire : Karyopherin-beta2 bound to hnRNP H2 PY-NLS
| Entire | Name: Karyopherin-beta2 bound to hnRNP H2 PY-NLS |
|---|---|
| Components |
|
-Supramolecule #1: Karyopherin-beta2 bound to hnRNP H2 PY-NLS
| Supramolecule | Name: Karyopherin-beta2 bound to hnRNP H2 PY-NLS / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transportin-1
| Macromolecule | Name: Transportin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 101.738812 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: GGSKMEYEWK PDEQGLQQIL QLLKESQSPD TTIQRTVQQK LEQLNQYPDF NNYLIFVLTK LKSEDEPTRS LSGLILKNNV KAHFQNFPN GVTDFIKSEC LNNIGDSSPL IRATVGILIT TIASKGELQN WPDLLPKLCS LLDSEDYNTC EGAFGALQKI C EDSAEILD ...String: GGSKMEYEWK PDEQGLQQIL QLLKESQSPD TTIQRTVQQK LEQLNQYPDF NNYLIFVLTK LKSEDEPTRS LSGLILKNNV KAHFQNFPN GVTDFIKSEC LNNIGDSSPL IRATVGILIT TIASKGELQN WPDLLPKLCS LLDSEDYNTC EGAFGALQKI C EDSAEILD SDVLDRPLNI MIPKFLQFFK HSSPKIRSHA VACVNQFIIS RTQALMLHID SFIENLFALA GDEEPEVRKN VC RALVMLL EVRMDRLLPH MHNIVEYMLQ RTQDQDENVA LEACEFWLTL AEQPICKDVL VRHLPKLIPV LVNGMKYSDI DII LLKGDV EEDETIPDSE QDIRPRFHRS RTVAQQHDED GIEEEDDDDD EIDDDDTISD WNLRKCSAAA LDVLANVYRD ELLP HILPL LKELLFHHEW VVKESGILVL GAIAEGCMQG MIPYLPELIP HLIQCLSDKK ALVRSITCWT LSRYAHWVVS QPPDT YLKP LMTELLKRIL DSNKRVQEAA CSAFATLEEE ACTELVPYLA YILDTLVFAF SKYQHKNLLI LYDAIGTLAD SVGHHL NKP EYIQMLMPPL IQKWNMLKDE DKDLFPLLEC LSSVATALQS GFLPYCEPVY QRCVNLVQKT LAQAMLNNAQ PDQYEAP DK DFMIVALDLL SGLAEGLGGN IEQLVARSNI LTLMYQCMQD KMPEVRQSSF ALLGDLTKAC FQHVKPCIAD FMPILGTN L NPEFISVCNN ATWAIGEISI QMGIEMQPYI PMVLHQLVEI INRPNTPKTL LENTAITIGR LGYVCPQEVA PMLQQFIRP WCTSLRNIRD NEEKDSAFRG ICTMISVNPS GVIQDFIFFC DAVASWINPK DDLRDMFCKI LHGFKNQVGD ENWRRFSDQF PLPLKERLA AFYGV UniProtKB: Transportin-1 |
-Macromolecule #2: Heterogeneous nuclear ribonucleoprotein H2, N-terminally processed
| Macromolecule | Name: Heterogeneous nuclear ribonucleoprotein H2, N-terminally processed type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.993811 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: GGSNSPDTAN DGFVRLRGLP FGCSKEEIVQ FFSGLEIVPN GMTLPVDFQG RSTGEAFVQF ASQEIAEKAL KKHKERIGHR YIEIFKSSR AEVRTHYDPP RKLMAMQRPG PYDRPGAGRG YNSIGRG UniProtKB: Heterogeneous nuclear ribonucleoprotein H2 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.7 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM Tris-HCl pH 7.5, 150 nM NaCl, 2 mM BME, 0.003125% [w/v] NP-40 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final 3D classification | Number classes: 7 / Software - Name: cryoSPARC |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.17 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Details: Non-uniform refinement / Number images used: 208572 |
-Atomic model buiding 1
| Details | Initial docking performed using Chimera, then manual model building by Coot, ISOLDE in ChimerX and Phenix real-space refinement. |
|---|---|
| Output model |  PDB-8sgh: |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X