[English] 日本語
 Yorodumi
Yorodumi- EMDB-40049: empty HBV Cp183 capsid with importin-beta, subparticle reconstruc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | empty HBV Cp183 capsid with importin-beta, subparticle reconstruction at 2fold location | |||||||||
 Map data Map data | empty HBV Cp183 capsid with importin-beta, subparticle reconstruction at 2fold location | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HBV / Cp183 / Importin-beta /  VIRAL PROTEIN VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule-dependent intracellular transport of viral material towards nucleus / T=4 icosahedral viral capsid / viral penetration into host nucleus / host cell cytoplasm / symbiont entry into host cell / structural molecule activity /  DNA binding / DNA binding /  RNA binding RNA bindingSimilarity search - Function | |||||||||
| Biological species |    Hepatitis B virus Hepatitis B virus | |||||||||
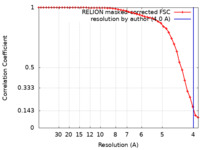
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.0 Å cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Kim C / Schlicksup CJ / Hadden-Perilla JA / Wang JC-Y / Zlotnick A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2023 Journal: J Biol Chem / Year: 2023Title: Structure of the Hepatitis B virus capsid quasi-6-fold with a trapped C-terminal domain reveals capsid movements associated with domain exit. Authors: Christine Kim / Christopher J Schlicksup / Carolina Pérez-Segura / Jodi A Hadden-Perilla / Joseph Che-Yen Wang / Adam Zlotnick /  Abstract: Many viruses undergo transient conformational change to surveil their environments for receptors and host factors. In Hepatitis B virus (HBV) infection, after the virus enters the cell, it is ...Many viruses undergo transient conformational change to surveil their environments for receptors and host factors. In Hepatitis B virus (HBV) infection, after the virus enters the cell, it is transported to the nucleus by interaction of the HBV capsid with an importin α/β complex. The interaction between virus and importins is mediated by nuclear localization signals on the capsid protein's C-terminal domain (CTD). However, CTDs are located inside the capsid. In this study, we asked where does a CTD exit the capsid, are all quasi-equivalent CTDs created equal, and does the capsid structure deform to facilitate CTD egress from the capsid? Here, we used Impβ as a tool to trap transiently exposed CTDs and examined this complex by cryo-electron microscopy. We examined an asymmetric reconstruction of a T = 4 icosahedral capsid and a focused reconstruction of a quasi-6-fold vertex (3.8 and 4.0 Å resolution, respectively). Both approaches showed that a subset of CTDs extended through a pore in the center of the quasi-6-fold complex. CTD egress was accompanied by enlargement of the pore and subtle changes in quaternary and tertiary structure of the quasi-6-fold. When compared to molecular dynamics simulations, structural changes were within the normal range of capsid flexibility. Although pore diameter was enlarged in the Impβ-bound reconstruction, simulations indicate that CTD egress does not exclusively depend on enlarged pores. In summary, we find that HBV surveillance of its environment by transient exposure of its CTD requires only modest conformational change of the capsid. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40049.map.gz emd_40049.map.gz | 6.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40049-v30.xml emd-40049-v30.xml emd-40049.xml emd-40049.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40049_fsc.xml emd_40049_fsc.xml | 4.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_40049.png emd_40049.png | 279.2 KB | ||
| Others |  emd_40049_half_map_1.map.gz emd_40049_half_map_1.map.gz emd_40049_half_map_2.map.gz emd_40049_half_map_2.map.gz | 4.9 MB 4.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40049 http://ftp.pdbj.org/pub/emdb/structures/EMD-40049 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40049 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40049 | HTTPS FTP |
-Related structure data
| Related structure data |  8ghsMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40049.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40049.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | empty HBV Cp183 capsid with importin-beta, subparticle reconstruction at 2fold location | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.94 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map 1
| File | emd_40049_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
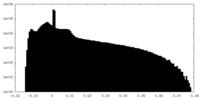
| Projections & Slices |
| ||||||||||||
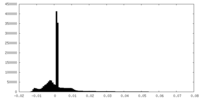
| Density Histograms |
-Half map: half map 2
| File | emd_40049_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
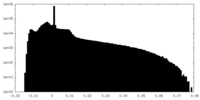
| Projections & Slices |
| ||||||||||||
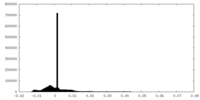
| Density Histograms |
- Sample components
Sample components
-Entire : Reassembled HBV empty Cp183 capsid with importin-beta
| Entire | Name: Reassembled HBV empty Cp183 capsid with importin-beta |
|---|---|
| Components |
|
-Supramolecule #1: Reassembled HBV empty Cp183 capsid with importin-beta
| Supramolecule | Name: Reassembled HBV empty Cp183 capsid with importin-beta / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Core protein Cp183 expressed in E coli |
|---|---|
| Source (natural) | Organism:    Hepatitis B virus Hepatitis B virus |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Hepatitis B virus Hepatitis B virus |
| Molecular weight | Theoretical: 16.840186 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MDIDPYKEFG ATVELLSFLP SDFFPSVRDL LDTAAALYRD ALESPEHCSP HHTALRQAIL CWGDLMTLAT WVGTNLEDPA SRDLVVSYV NTNVGLKFRQ LLWFHISCLT FGRETVLEYL VSFGVWIRTP PAYRPPNAPI LSTLPETAAA AA UniProtKB:  Capsid protein Capsid protein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 0.15 M NaCl, 20 mM Tris-HCl pH 7.5, and 10 mM DTT |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
| Details | Empty HBV capsid : Importin-beta sample at 1:205 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Details | A hexamer model was flexibly fitted into the hexamer maps using Alphafold, ISOLDE, PHENIX, and Coot. |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-8ghs: |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X