[English] 日本語
 Yorodumi
Yorodumi- PDB-3jcl: Cryo-electron microscopy structure of a coronavirus spike glycopr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3jcl | ||||||
|---|---|---|---|---|---|---|---|
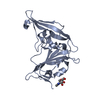
| Title | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer | ||||||
 Components Components | Spike glycoprotein Spike protein Spike protein | ||||||
 Keywords Keywords |  VIRAL PROTEIN / VIRAL PROTEIN /  Coronavirus / viral fusion proteins / Coronavirus / viral fusion proteins /  viral spike / viral spike /  peplomer peplomer | ||||||
| Function / homology |  Function and homology information Function and homology informationendocytosis involved in viral entry into host cell / host cell Golgi apparatus / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / host cell plasma membrane / virion membrane / viral envelope / host cell plasma membrane / virion membrane /  membrane / identical protein binding membrane / identical protein bindingSimilarity search - Function | ||||||
| Biological species |   Murine hepatitis virus strain A59 Murine hepatitis virus strain A59 | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4 Å cryo EM / Resolution: 4 Å | ||||||
 Authors Authors | Walls, A.C. / Tortorici, M.A. / Bosch, B.J. / Frenz, B. / Rottier, P.J.M. / DiMaio, F. / Rey, F.A. / Veesler, D. | ||||||
 Citation Citation |  Journal: Nature / Year: 2016 Journal: Nature / Year: 2016Title: Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Authors: Alexandra C Walls / M Alejandra Tortorici / Berend-Jan Bosch / Brandon Frenz / Peter J M Rottier / Frank DiMaio / Félix A Rey / David Veesler /    Abstract: The tremendous pandemic potential of coronaviruses was demonstrated twice in the past few decades by two global outbreaks of deadly pneumonia. Entry of coronaviruses into cells is mediated by the ...The tremendous pandemic potential of coronaviruses was demonstrated twice in the past few decades by two global outbreaks of deadly pneumonia. Entry of coronaviruses into cells is mediated by the transmembrane spike glycoprotein S, which forms a trimer carrying receptor-binding and membrane fusion functions. S also contains the principal antigenic determinants and is the target of neutralizing antibodies. Here we present the structure of a mouse coronavirus S trimer ectodomain determined at 4.0 Å resolution by single particle cryo-electron microscopy. It reveals the metastable pre-fusion architecture of S and highlights key interactions stabilizing it. The structure shares a common core with paramyxovirus F proteins, implicating mechanistic similarities and an evolutionary connection between these viral fusion proteins. The accessibility of the highly conserved fusion peptide at the periphery of the trimer indicates potential vaccinology strategies to elicit broadly neutralizing antibodies against coronaviruses. Finally, comparison with crystal structures of human coronavirus S domains allows rationalization of the molecular basis for species specificity based on the use of spatially contiguous but distinct domains. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3jcl.cif.gz 3jcl.cif.gz | 624 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3jcl.ent.gz pdb3jcl.ent.gz | 512.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3jcl.json.gz 3jcl.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jc/3jcl https://data.pdbj.org/pub/pdb/validation_reports/jc/3jcl ftp://data.pdbj.org/pub/pdb/validation_reports/jc/3jcl ftp://data.pdbj.org/pub/pdb/validation_reports/jc/3jcl | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6526MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein |  Spike protein / S glycoprotein / E2 / Peplomer protein / Spike protein S1 / 90B / Spike protein S2 / 90A Spike protein / S glycoprotein / E2 / Peplomer protein / Spike protein S1 / 90B / Spike protein S2 / 90AMass: 139728.984 Da / Num. of mol.: 3 / Fragment: UNP residues 15-1231 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Murine hepatitis virus strain A59 / Strain: A59 / Gene: 3, S / Production host: Murine hepatitis virus strain A59 / Strain: A59 / Gene: 3, S / Production host:   Drosophila melanogaster (fruit fly) / Strain (production host): S2 / References: UniProt: P11224 Drosophila melanogaster (fruit fly) / Strain (production host): S2 / References: UniProt: P11224 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Murine hepatitis virus protein S / Type: VIRUS / Details: Trimer |
|---|---|
| Molecular weight | Value: 0.42 MDa / Experimental value: NO |
| Buffer solution | Name: 20 mM Tris-HCl, 100 mM NaCl / pH: 7.5 / Details: 20 mM Tris-HCl, 100 mM NaCl |
| Specimen | Conc.: 1.85 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Instrument: GATAN CRYOPLUNGE 3 / Cryogen name: ETHANE / Temp: 93 K / Humidity: 90 % Details: Blot for 3.5 seconds before plunging into liquid ethane (GATAN CRYOPLUNGE 3). Method: Blot for 3.5 seconds before plunging |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS / Date: Dec 9, 2014 |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 22500 X / Calibrated magnification: 38022 X / Nominal defocus max: 5000 nm / Nominal defocus min: 2000 nm / Cs Bright-field microscopy / Nominal magnification: 22500 X / Calibrated magnification: 38022 X / Nominal defocus max: 5000 nm / Nominal defocus min: 2000 nm / Cs : 2.7 mm : 2.7 mm |
| Specimen holder | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 53 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
| Image scans | Num. digital images: 1600 |
- Processing
Processing
| EM software |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Details: Each particle | |||||||||||||||
| Symmetry | Point symmetry : C3 (3 fold cyclic : C3 (3 fold cyclic ) ) | |||||||||||||||
3D reconstruction | Method: Projection-matching / Resolution: 4 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 82000 / Nominal pixel size: 1.46 Å / Actual pixel size: 1.46 Å / Details: (Single particle--Applied symmetry: C3) / Symmetry type: POINT | |||||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT / Space: REAL / Details: REFINEMENT PROTOCOL--flexible | |||||||||||||||
| Atomic model building |
| |||||||||||||||
| Refinement step | Cycle: LAST
|
 Movie
Movie Controller
Controller










 PDBj
PDBj