[English] 日本語
 Yorodumi
Yorodumi- EMDB-35691: Photochromobilin-free form of Arabidopsis thaliana phytochrome A ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Photochromobilin-free form of Arabidopsis thaliana phytochrome A - apo-AtphyA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phytochrome A / Photochromobilin-free form / far-red light receptor /  PLANT PROTEIN PLANT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to continuous far red light stimulus by the high-irradiance response system / response to very low fluence red light stimulus / far-red light photoreceptor activity / red light signaling pathway / red or far-red light photoreceptor activity /  gravitropism / response to far red light / gravitropism / response to far red light /  phototropism / phototropism /  photomorphogenesis / detection of visible light ...response to continuous far red light stimulus by the high-irradiance response system / response to very low fluence red light stimulus / far-red light photoreceptor activity / red light signaling pathway / red or far-red light photoreceptor activity / photomorphogenesis / detection of visible light ...response to continuous far red light stimulus by the high-irradiance response system / response to very low fluence red light stimulus / far-red light photoreceptor activity / red light signaling pathway / red or far-red light photoreceptor activity /  gravitropism / response to far red light / gravitropism / response to far red light /  phototropism / phototropism /  photomorphogenesis / detection of visible light / response to arsenic-containing substance / phosphorelay sensor kinase activity / response to cold / negative regulation of translation / photomorphogenesis / detection of visible light / response to arsenic-containing substance / phosphorelay sensor kinase activity / response to cold / negative regulation of translation /  nuclear body / nuclear body /  protein kinase activity / nuclear speck / protein kinase activity / nuclear speck /  mRNA binding / regulation of DNA-templated transcription / protein homodimerization activity / identical protein binding / mRNA binding / regulation of DNA-templated transcription / protein homodimerization activity / identical protein binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.77 Å cryo EM / Resolution: 3.77 Å | |||||||||
 Authors Authors | Zhang Y / Ma C / Zhao J / Gao N / Wang J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Structural insights into plant phytochrome A as a highly sensitized photoreceptor. Authors: Yuxuan Zhang / Xiaoli Lin / Chengying Ma / Jun Zhao / Xiaojin Shang / Zhengdong Wang / Bin Xu / Ning Gao / Xing Wang Deng / Jizong Wang /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35691.map.gz emd_35691.map.gz | 3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35691-v30.xml emd-35691-v30.xml emd-35691.xml emd-35691.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35691.png emd_35691.png | 33.1 KB | ||
| Filedesc metadata |  emd-35691.cif.gz emd-35691.cif.gz | 6 KB | ||
| Others |  emd_35691_half_map_1.map.gz emd_35691_half_map_1.map.gz emd_35691_half_map_2.map.gz emd_35691_half_map_2.map.gz | 45.2 MB 45.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35691 http://ftp.pdbj.org/pub/emdb/structures/EMD-35691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35691 | HTTPS FTP |
-Related structure data
| Related structure data |  8isiMC  8isjC  8iskC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35691.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35691.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35691_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
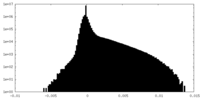
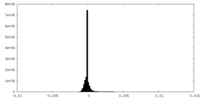
| Density Histograms |
-Half map: #2
| File | emd_35691_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
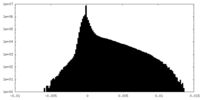
| Density Histograms |
- Sample components
Sample components
-Entire : Photochromobilin-free form of Arabidopsis thaliana phytochrome A ...
| Entire | Name: Photochromobilin-free form of Arabidopsis thaliana phytochrome A (apo-AtphyA) |
|---|---|
| Components |
|
-Supramolecule #1: Photochromobilin-free form of Arabidopsis thaliana phytochrome A ...
| Supramolecule | Name: Photochromobilin-free form of Arabidopsis thaliana phytochrome A (apo-AtphyA) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
-Macromolecule #1: Phytochrome A
| Macromolecule | Name: Phytochrome A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
| Molecular weight | Theoretical: 125.17143 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: MEKKMSGSRP TQSSEGSRRS RHSARIIAQT TVDAKLHADF EESGSSFDYS TSVRVTGPVV ENQPPRSDKV TTTYLHHIQK GKLIQPFGC LLALDEKTFK VIAYSENASE LLTMASHAVP SVGEHPVLGI GTDIRSLFTA PSASALQKAL GFGDVSLLNP I LVHCRTSA ...String: MEKKMSGSRP TQSSEGSRRS RHSARIIAQT TVDAKLHADF EESGSSFDYS TSVRVTGPVV ENQPPRSDKV TTTYLHHIQK GKLIQPFGC LLALDEKTFK VIAYSENASE LLTMASHAVP SVGEHPVLGI GTDIRSLFTA PSASALQKAL GFGDVSLLNP I LVHCRTSA KPFYAIIHRV TGSIIIDFEP VKPYEVPMTA AGALQSYKLA AKAITRLQSL PSGSMERLCD TMVQEVFELT GY DRVMAYK FHEDDHGEVV SEVTKPGLEP YLGLHYPATD IPQAARFLFM KNKVRMIVDC NAKHARVLQD EKLSFDLTLC GST LRAPHS CHLQYMANMD SIASLVMAVV VNEEDGEGDA PDATTQPQKR KRLWGLVVCH NTTPRFVPFP LRYACEFLAQ VFAI HVNKE VELDNQMVEK NILRTQTLLC DMLMRDAPLG IVSQSPNIMD LVKCDGAALL YKDKIWKLGT TPSEFHLQEI ASWLC EYHM DSTGLSTDSL HDAGFPRALS LGDSVCGMAA VRISSKDMIF WFRSHTAGEV RWGGAKHDPD DRDDARRMHP RSSFKA FLE VVKTRSLPWK DYEMDAIHSL QLILRNAFKD SETTDVNTKV IYSKLNDLKI DGIQELEAVT SEMVRLIETA TVPILAV DS DGLVNGWNTK IAELTGLSVD EAIGKHFLTL VEDSSVEIVK RMLENALEGT EEQNVQFEIK THLSRADAGP ISLVVNAC A SRDLHENVVG VCFVAHDLTG QKTVMDKFTR IEGDYKAIIQ NPNPLIPPIF GTDEFGWCTE WNPAMSKLTG LKREEVIDK MLLGEVFGTQ KSCCRLKNQE AFVNLGIVLN NAVTSQDPEK VSFAFFTRGG KYVECLLCVS KKLDREGVVT GVFCFLQLAS HELQQALHV QRLAERTAVK RLKALAYIKR QIRNPLSGIM FTRKMIEGTE LGPEQRRILQ TSALCQKQLS KILDDSDLES I IEGCLDLE MKEFTLNEVL TASTSQVMMK SNGKSVRITN ETGEEVMSDT LYGDSIRLQQ VLADFMLMAV NFTPSGGQLT VS ASLRKDQ LGRSVHLANL EIRLTHTGAG IPEFLLNQMF GTEEDVSEEG LSLMVSRKLV KLMNGDVQYL RQAGKSSFII TAE LAAANK UniProtKB: Phytochrome A |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.77 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 873442 |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X