+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
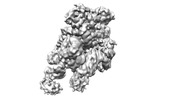
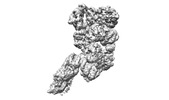
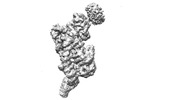
| Title | base module state 1 of Tetrahymena IFT-A | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  intraflagellar transport complex / intraflagellar transport complex /  PROTEIN TRANSPORT PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.6 Å cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Ma Y / Wu J / Lei M | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train. Authors: Yuanyuan Ma / Jun He / Shaobai Li / Deqiang Yao / Chenhui Huang / Jian Wu / Ming Lei /  Abstract: Intraflagellar transport (IFT) trains, the polymers composed of two multi-subunit complexes, IFT-A and IFT-B, carry out bidirectional intracellular transport in cilia, vital for cilia biogenesis and ...Intraflagellar transport (IFT) trains, the polymers composed of two multi-subunit complexes, IFT-A and IFT-B, carry out bidirectional intracellular transport in cilia, vital for cilia biogenesis and signaling. IFT-A plays crucial roles in the ciliary import of membrane proteins and the retrograde cargo trafficking. However, the molecular architecture of IFT-A and the assembly mechanism of the IFT-A into the IFT trains in vivo remains elusive. Here, we report the cryo-electron microscopic structures of the IFT-A complex from protozoa Tetrahymena thermophila. We find that IFT-A complexes present two distinct, elongated and folded states. Remarkably, comparison with the in situ cryo-electron tomography structure of the anterograde IFT train unveils a series of adjustments of the flexible arms in apo IFT-A when incorporated into the anterograde train. Our results provide an atomic-resolution model for the IFT-A complex and valuable insights into the assembly mechanism of anterograde IFT trains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34895.map.gz emd_34895.map.gz | 135.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34895-v30.xml emd-34895-v30.xml emd-34895.xml emd-34895.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34895.png emd_34895.png | 57.3 KB | ||
| Others |  emd_34895_half_map_1.map.gz emd_34895_half_map_1.map.gz emd_34895_half_map_2.map.gz emd_34895_half_map_2.map.gz | 114.1 MB 114.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34895 http://ftp.pdbj.org/pub/emdb/structures/EMD-34895 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34895 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34895 | HTTPS FTP |
-Related structure data
| Related structure data |  8hmcMC  8hmdC  8hmeC  8hmfC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34895.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34895.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34895_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34895_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : IFT-A_base-1
| Entire | Name: IFT-A_base-1 |
|---|---|
| Components |
|
-Supramolecule #1: IFT-A_base-1
| Supramolecule | Name: IFT-A_base-1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
-Macromolecule #1: Intraflagellar transport protein 122 homolog
| Macromolecule | Name: Intraflagellar transport protein 122 homolog / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
| Molecular weight | Theoretical: 143.939422 KDa |
| Sequence | String: MKCSELWTEI IPKNDPNSNI TNQVWSCSFN NDGSILLACV GDAILVYDSF TGNLVNKAIR GGQKEQINCI KFSKDGKRFA TASNDKTVW VWSFDVNKNP KLAPEVKYSH TDKVLCLAFN PLTHQIFSGG AQDYALWTPD QQSIEKSKFK DKIITCAWSW D GLYLGFGT ...String: MKCSELWTEI IPKNDPNSNI TNQVWSCSFN NDGSILLACV GDAILVYDSF TGNLVNKAIR GGQKEQINCI KFSKDGKRFA TASNDKTVW VWSFDVNKNP KLAPEVKYSH TDKVLCLAFN PLTHQIFSGG AQDYALWTPD QQSIEKSKFK DKIITCAWSW D GLYLGFGT MGGIIGIRNR ALEQLAEIQR SFPIWCMDWT PITPDYQNSV LVVGVWEQSI AHYNIQGQQL GSDKKLTYDP ID INFSTQG EFLLVSGTNN KCTLYTREGG FLIDVATKND WVWNNKLKPV IKEAGSVAVQ DQLHVALTTN DGEISMFKIQ QLT VHALEG DKYAYRDNLT DIVVQSMSSN QRIRLKCKEL VKNISIYKDK LAAHLTERIL IYGTSSEDSQ MKYKLHKRIV KKVE AHILE IVSNHLIACV KNKVQLYQLS GDLEKEWVFD SRVNCIKIIG GMPNKEGAYV GLKNGQIFKI FVDNSFPIPI LNHNI SIKN IDASQKRKRL AVVDKNSNLT VYDLSNKEQV FQEMNIVSCA FNNNFDDLLA FSSTDTTFIR CSNHPPLSQK ISGQVV GFK ANKLFVLNDN VMSTIDVSMT QTMIKYLERK EFQSAYEISL LGVTDQDFLY LGNEALIATQ FAIARKCYTR IKKLDFI YL LEKAEKDFKK NTFVEGFYQG EIYALQTKFY EAENALTKSN LASQAKEMWI ILKEFDKAMR ITDTGASPIN RGQNKEIN S DASRKDLLIQ RAEWEVKQGN WKKGGELYIQ AEVYKKAIEV YVTNNFSEGI VEVCRNADAE TQRKEIEQCA AYFKKAKLH QYTKEAYLKL QDNKALLQLN IDLEKWDEAM TLAKAHPEFM EIVKLPYANW LAKQDRYEES LKAYRKIGKH DMATKMLYNL SQNAVFEKR FSDAALFTWM IATEHLGLIR NMKAPTPEDI ENLMKFYQYR DDAEIYFAYS KVQSFVDEPF LPLSGHAYML N IFNAARFA INKLGNRQLY GVQHSYLYYS LGKVSKQLEG YKTARICYEK LASYKIPTEW SEEIELSTLL IRSKPYSDQE SL LPICNRC YNQNPALTEG NRCSSCMHQF QNCFISFENL PLVEFKLDPK ITHKRFIELI NSDKSKTQPN KKKKKANDGW QES HQGDQQ VLTFNNSPQK GGNDNESSPF LDKLNEVFEI QQTTQEYIPV TLDENIVSSL SIDEVYWVDY TKYCYTAEIK YYKS MLTDI PLKNCQECGT FYILDEYEFE ISKTQKCPFC RSVDERAGPQ KDVFDF |
-Macromolecule #2: WD40 repeat protein
| Macromolecule | Name: WD40 repeat protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
| Molecular weight | Theoretical: 138.076984 KDa |
| Sequence | String: MFVFLSKKIG MPNNAKIEAV CWNKEQGWIA AGGEKGILKV LKIEDQRQKD GSNPTTTGQL LVNNTLEYHT SNVYLVTWND RYRKLTSVE ENGIIVVWAY FRELQLWKEE MINDRKKSVV RDLKWSPDGQ KVCIAYEDGA VIVGSVEGSP LWRKEFPHKL A LIEWSPDS ...String: MFVFLSKKIG MPNNAKIEAV CWNKEQGWIA AGGEKGILKV LKIEDQRQKD GSNPTTTGQL LVNNTLEYHT SNVYLVTWND RYRKLTSVE ENGIIVVWAY FRELQLWKEE MINDRKKSVV RDLKWSPDGQ KVCIAYEDGA VIVGSVEGSP LWRKEFPHKL A LIEWSPDS KMMIFGTPEG EVRVYDSQGM EMQRLKIHCL QKLIDPSKSF SPDLPLAAID WFPQAKMYTD DTPPGLCIAY QC GRVQLMK NEKDDEPVLI DTMMKIFTVK WNPSGSMFAI SGTQDENGEL KGVVQFYSNA GHHLKTLRVP GSDRVTGVSW EGS GLRIAM AVNQAIFFAN IKPDHKWAYM SDGTLAFAYQ KIDRVEYTII FWDTINNKKN LKFVKNLCCL RASGDLCCIV TELI GYDLW QIDLCNSIGS PIDSKQINIY PNAVTMTKTH IIVCSQDHVY AWQYKNQVER LTTFEQQTGL RRVGREQAWF IDKEN DSNN QYDKDTYDVE PQSEDVICTV AANENFLLVA RISGTINVYT FPHISLENKL HIQTRPSQMS LNKDATRLAI IDHNGN LNI LKITPQGDEL LPFEKKECWF VKYSDDIPES FVFMERSRMY IVNDQTPEEP IITEGYICQY KDFQVKIVYL DDIMKSP DG VLRKEELTLE IEANILKDIK DQLYKKSMQE MFIVIKQHDN DCLWRVYAKK ALEDNDFDSA ENAFVQCKDY ASLKFLQR V RELDDRERQR AEIQCYFNKV EEAEEIYNKI ERRDLSIQMR MKLGDWAQVV DQIREGTGQD VELQKARLEL GNYYAENFQ WDKAAKQYAL AKYNPGLIEA YTRIQDYEGL EKLIAEIPER NELLQDMGDR FQQAGLCDAA VKCYEKFGDI KQAIDCCVLL NHWNLAVEL AEQYNFVQIE GLLVQYANQL LEKRRKLEAA ELYRKAKRNT EAAKILSQIA DDLTERDANP LNIKKMYVMA A LEVDLYKK RMLDATMTGQ ATSTAKTLNS LITSTINTSS ADKILNNPWR GAEAWHFFIL AQRQLYNGQF KYALKSALRL GE YELEIDQ KKIYSLIAIA AYYNKSFREC SRAFVKLQNL ENITEDEKER YEAIAVSIFT KHPPLDSPCE YTPCVGKNCS QQV SEYDIH CRACGSNFSP CVASGRPIFQ KEFYQCKNCR HKMIESEVSR LKLYNCALCH SPIDFKRFTE SNNNNNNNI |
-Macromolecule #3: Tetratricopeptide repeat protein
| Macromolecule | Name: Tetratricopeptide repeat protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
| Molecular weight | Theoretical: 154.216141 KDa |
| Sequence | String: MQNQQQKMQT ILIAQSQVYY YIREGFWSTM QRFCQEQYKA FGDPFFIFWK AYGLYQEGLP NEAINELTSI QHKKEIQYAT IVALITYHL STNIVDRETV QNLKFEESTQ RRLSSDKAIC LAAFFYAFNK EHAKARDLID EIHSDNFNIR IASAWCYLLE G GKFLEKSV ...String: MQNQQQKMQT ILIAQSQVYY YIREGFWSTM QRFCQEQYKA FGDPFFIFWK AYGLYQEGLP NEAINELTSI QHKKEIQYAT IVALITYHL STNIVDRETV QNLKFEESTQ RRLSSDKAIC LAAFFYAFNK EHAKARDLID EIHSDNFNIR IASAWCYLLE G GKFLEKSV QLFEELYNEQ HEINKNLESL MGRSKANEMI KKFDISLNTI NEINVLYPDF KGGLIEKAKL LMTVDDWEQL VD YCNKILY DDDKNIMALM LLTFYTFARE GDIETGCERL QKLIQAVEFS ESRNMQLMFK ISQVFSRISG RNTQILKFTM KLV NQCKQL SPLNAQYFCE LAQQLLMVNQ FERAEQYFQE ASAIDVDNKE CLMGLILSKI MQGQTEDAES QIDFINQTTN NGER TSEIA YLEALVSTKQ ENVDPRVTIK LLEESLKLHI AQANRLYPSF DFYIVLNPDF LMSLSQAYFF QVGMKEMLAG KQPQN GVAS KGTKLLDFII KKIPGLIPAY LLQAKGKMSM GNTQEALKSV TKVIEQDPKN EEAYILSAMI ASSSKNFSLA QNQLQQ ALS NNFMIRDNPL FMLVKGEVEY AQGSYQACLE TMKAAYEIPE VKDKANQSKV VSAMSVLQFS DKDRCSIFLL YAKALQQ NN NSKEAKKIMT QAISQFTGTT EEVNVLIANS EIALQSGDVK KAISILKGVP QESPYFLRAR QILADVYLDQ LRDRRNYA K CYADLIEIDP SFDNYKMLGD ALMKIREPEE ASRAYEKAAL IKPDDEQIIQ LLGLSLCQTH DYNKALTYYE NALRMNPKR LDLIIDLGKL CIQIKNFNRA EEILKPDIFS DEYQLPTYQN LKRNQEGFYL IAKLNIKRTP PGVFTPIDMY RKALKKSIQI QIDVIEKAK QEGEDVEKER KTLADMYIEL AKYTNQYEKN EKATLDILAE ASKYTNNQDT MSKTVGNQEK ILELEVQMYF K SNQKLECE NKCNLLLKLN PNNDLACLTL AELLLQKDEY SQAIEQFKKI LQDRPNNYGI LAKLIDFFRR SFQINEAKTY IE RAEKKAT NTNDPGLCYC RGLYHKYNRS PKDALNEFSK AKKSSQYAEE SLVNMIDIYL NPDQDLYYSN VEEGPKVVDE VNL RACESL LREMQIRASY LRYIVMESYV FFLGGPRYKG GLEQGLKNLN DILKTNNDYI PAMLALAVGK FIQKKSTDAK NLLK LLWKR QYTTEYGEDL ERAWLLSADS FIAIQKYDSA EEILKKCLKY NQSCGKAEEY MGLIKEKEQS YVDAATHYEK AYKLT NEKS ASIAFRLSFN YLKAKRYVDC INICKKILVL FPNYPKIEKD CLEKARQALK |
-Macromolecule #4: Intraflagellar transport protein 43 homolog
| Macromolecule | Name: Intraflagellar transport protein 43 homolog / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
| Molecular weight | Theoretical: 16.711193 KDa |
| Sequence | String: MAAKGKQGWG FGGKDQNVKI DTSQQDQKKQ NIWEQNNEDL IFVPDLTQEA QEQEVSKVSA PPNQPTVQVQ DINELQKFTK INTLPQTEE GVDLSQLMQI LSPVEDIKEK DEAWEFLQLK TQIYEIVSNM YGGNELIDDD DEDDENQ |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 538078 |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X

















