[English] 日本語
 Yorodumi
Yorodumi- EMDB-34094: Structural basis of vitamin C recognition and transport by mammal... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationVitamin C (ascorbate) metabolism / L-ascorbate:sodium symporter activity / L-ascorbic acid transmembrane transporter activity / L-ascorbic acid transmembrane transport / dehydroascorbic acid transmembrane transporter activity / dehydroascorbic acid transport / sodium ion transmembrane transporter activity / intracellular organelle / sodium ion transport /  brush border ...Vitamin C (ascorbate) metabolism / L-ascorbate:sodium symporter activity / L-ascorbic acid transmembrane transporter activity / L-ascorbic acid transmembrane transport / dehydroascorbic acid transmembrane transporter activity / dehydroascorbic acid transport / sodium ion transmembrane transporter activity / intracellular organelle / sodium ion transport / brush border ...Vitamin C (ascorbate) metabolism / L-ascorbate:sodium symporter activity / L-ascorbic acid transmembrane transporter activity / L-ascorbic acid transmembrane transport / dehydroascorbic acid transmembrane transporter activity / dehydroascorbic acid transport / sodium ion transmembrane transporter activity / intracellular organelle / sodium ion transport /  brush border / basal plasma membrane / lung development / brush border / basal plasma membrane / lung development /  brain development / response to toxic substance / apical plasma membrane / brain development / response to toxic substance / apical plasma membrane /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | ||||||||||||||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | ||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | ||||||||||||||||||
 Authors Authors | She J / Wang M / He J / Zhang K / Li S | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter. Authors: Mingxing Wang / Jin He / Shanshan Li / Qianwen Cai / Kaiming Zhang / Ji She /  Abstract: Vitamin C (L-ascorbic acid) is an essential nutrient for human health, and its deficiency has long been known to cause scurvy. Sodium-dependent vitamin C transporters (SVCTs) are responsible for ...Vitamin C (L-ascorbic acid) is an essential nutrient for human health, and its deficiency has long been known to cause scurvy. Sodium-dependent vitamin C transporters (SVCTs) are responsible for vitamin C uptake and tissue distribution in mammals. Here, we present cryogenic electron microscopy structures of mouse SVCT1 in both the apo and substrate-bound states. Mouse SVCT1 forms a homodimer with each protomer containing a core domain and a gate domain. The tightly packed extracellular interfaces between the core domain and gate domain stabilize the protein in an inward-open conformation for both the apo and substrate-bound structures. Vitamin C binds at the core domain of each subunit, and two potential sodium ions are identified near the binding site. The coordination of sodium ions by vitamin C explains their coupling transport. SVCTs probably deliver substrate through an elevator mechanism in combination with local structural arrangements. Altogether, our results reveal the molecular mechanism by which SVCTs recognize vitamin C and lay a foundation for further mechanistic studies on SVCT substrate transport. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34094.map.gz emd_34094.map.gz | 203.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34094-v30.xml emd-34094-v30.xml emd-34094.xml emd-34094.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34094.png emd_34094.png | 99.8 KB | ||
| Others |  emd_34094_additional_1.map.gz emd_34094_additional_1.map.gz emd_34094_half_map_1.map.gz emd_34094_half_map_1.map.gz emd_34094_half_map_2.map.gz emd_34094_half_map_2.map.gz | 111.4 MB 200.1 MB 200.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34094 http://ftp.pdbj.org/pub/emdb/structures/EMD-34094 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34094 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34094 | HTTPS FTP |
-Related structure data
| Related structure data |  7ytwMC  7ytyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34094.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34094.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
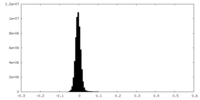
-Additional map: #1
| File | emd_34094_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
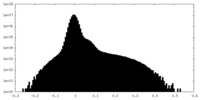
-Half map: #2
| File | emd_34094_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34094_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SVCT1
| Entire | Name: SVCT1 |
|---|---|
| Components |
|
-Supramolecule #1: SVCT1
| Supramolecule | Name: SVCT1 / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Macromolecule #1: Solute carrier family 23 member 1
| Macromolecule | Name: Solute carrier family 23 member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 65.600438 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKTPEDPGSP KQHEVVDSAG TSTRDRQAPL PTEPKFDMLY KIEDVPPWYL CILLGFQHYL TCFSGTIAVP FLLAEALCVG RDQHMVSQL IGTIFTCVGI TTLIQTTVGI RLPLFQASAF AFLVPAKSIL ALERWKCPSE EEIYGNWSMP LNTSHIWHPR I REVQGAIM ...String: MKTPEDPGSP KQHEVVDSAG TSTRDRQAPL PTEPKFDMLY KIEDVPPWYL CILLGFQHYL TCFSGTIAVP FLLAEALCVG RDQHMVSQL IGTIFTCVGI TTLIQTTVGI RLPLFQASAF AFLVPAKSIL ALERWKCPSE EEIYGNWSMP LNTSHIWHPR I REVQGAIM VSSMVEVVIG LMGLPGALLS YIGPLTVTPT VSLIGLSVFQ AAGDRAGSHW GISACSILLI VLFSQYLRNL TF LLPVYRW GKGLTLFRVQ IFKMFPIVLA IMTVWLLCYV LTLTDVLPAD PTVYGFQART DARGDIMAIS PWIRIPYPCQ WGL PTVTVA AVLGMFSATL AGIIESIGDY YACARLAGAP PPPVHAINRG IFTEGICCII AGLLGTGNGS TSSSPNIGVL GITK VGSRR VVQYGAGIML ILGAIGKFTA LFASLPDPIL GGMFCTLFGM ITAVGLSNLQ FVDMNSSRNL FVLGFSMFFG LTLPN YLDS NPGAINTGIP EVDQILTVLL TTEMFVGGCL AFILDNTVPG SPEERGLIQW KAGAHANSET SASLKSYDFP FGMGMV KRT TFFRYIPICP VFRGFSKKTQ NQPPVLEDTP DNIETGSVCT KV |
-Macromolecule #2: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 2 / Number of copies: 4 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #3: ASCORBIC ACID
| Macromolecule | Name: ASCORBIC ACID / type: ligand / ID: 3 / Number of copies: 2 / Formula: ASC |
|---|---|
| Molecular weight | Theoretical: 176.124 Da |
| Chemical component information | 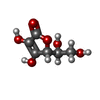 ChemComp-ASC: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 2370225 |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X