[English] 日本語
 Yorodumi
Yorodumi- EMDB-33886: PIC-Mediator (MED19-MED26 double mutant) in complex with +1 nucle... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PIC-Mediator (MED19-MED26 double mutant) in complex with +1 nucleosome (T40N) | |||||||||
 Map data Map data | PIC-Mediator (MED19-MED26 double mutant) in complex with plus 1 nucleosome (T40N) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Sus scrofa (pig) Sus scrofa (pig) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 14.85 Å cryo EM / Resolution: 14.85 Å | |||||||||
 Authors Authors | Chen X / Wang X / Liu W / Ren Y / Qu X / Li J / Yin X / Xu Y | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structures of +1 nucleosome-bound PIC-Mediator complex. Authors: Xizi Chen / Xinxin Wang / Weida Liu / Yulei Ren / Xuechun Qu / Jiabei Li / Xiaotong Yin / Yanhui Xu /  Abstract: RNA polymerase II-mediated eukaryotic transcription starts with the assembly of the preinitiation complex (PIC) on core promoters. The +1 nucleosome is well positioned about 40 base pairs downstream ...RNA polymerase II-mediated eukaryotic transcription starts with the assembly of the preinitiation complex (PIC) on core promoters. The +1 nucleosome is well positioned about 40 base pairs downstream of the transcription start site (TSS) and is commonly known as a barrier of transcription. The +1 nucleosome-bound PIC-Mediator structures show that PIC-Mediator prefers binding to T40N nucleosome located 40 base pairs downstream of TSS and contacts T50N but not the T70N nucleosome. The nucleosome facilitates the organization of PIC-Mediator on the promoter by binding TFIIH subunit p52 and Mediator subunits MED19 and MED26 and may contribute to transcription initiation. PIC-Mediator exhibits multiple nucleosome-binding patterns, supporting a structural role of the +1 nucleosome in the coordination of PIC-Mediator assembly. Our study reveals the molecular mechanism of PIC-Mediator organization on chromatin and underscores the significance of the +1 nucleosome in regulating transcription initiation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33886.map.gz emd_33886.map.gz | 79.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33886-v30.xml emd-33886-v30.xml emd-33886.xml emd-33886.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33886.png emd_33886.png | 29.5 KB | ||
| Others |  emd_33886_additional_1.map.gz emd_33886_additional_1.map.gz emd_33886_half_map_1.map.gz emd_33886_half_map_1.map.gz emd_33886_half_map_2.map.gz emd_33886_half_map_2.map.gz | 13.6 MB 90.2 MB 90.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33886 http://ftp.pdbj.org/pub/emdb/structures/EMD-33886 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33886 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33886 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33886.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33886.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PIC-Mediator (MED19-MED26 double mutant) in complex with plus 1 nucleosome (T40N) | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.89724 Å | ||||||||||||||||||||
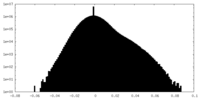
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Nucleosome of PIC-Mediator (MED19-MED26 double mutant) in complex...
| File | emd_33886_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nucleosome of PIC-Mediator (MED19-MED26 double mutant) in complex with plus 1 nucleosome (T40N) | ||||||||||||
| Projections & Slices |
| ||||||||||||
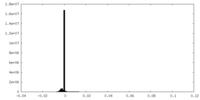
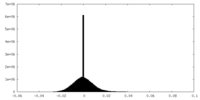
| Density Histograms |
-Half map: #1
| File | emd_33886_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
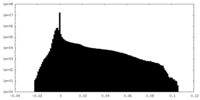
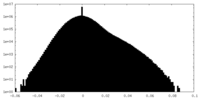
| Density Histograms |
-Half map: #2
| File | emd_33886_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
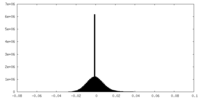
| Density Histograms |
- Sample components
Sample components
-Entire : PIC-Mediator (MED19-MED26 double mutant) in complex with +1 nucle...
| Entire | Name: PIC-Mediator (MED19-MED26 double mutant) in complex with +1 nucleosome (T40N) |
|---|---|
| Components |
|
-Supramolecule #1: PIC-Mediator (MED19-MED26 double mutant) in complex with +1 nucle...
| Supramolecule | Name: PIC-Mediator (MED19-MED26 double mutant) in complex with +1 nucleosome (T40N) type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Sus scrofa (pig) Sus scrofa (pig) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: OTHER |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 14.85 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 4189 |
 Movie
Movie Controller
Controller













 Z
Z Y
Y X
X