[English] 日本語
 Yorodumi
Yorodumi- EMDB-33503: Cryo-EM structure of the purinergic receptor P2Y1R in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the purinergic receptor P2Y1R in complex with 2MeSADP and G11 | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords |  G protein-coupled receptor / purinergic receptor / P2Y1R / G protein-coupled receptor / purinergic receptor / P2Y1R /  ligand binding / ligand binding /  signal transduction / signal transduction /  MEMBRANE PROTEIN MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled ATP receptor activity / relaxation of muscle /  A1 adenosine receptor binding / G protein-coupled ADP receptor activity / A1 adenosine receptor binding / G protein-coupled ADP receptor activity /  P2Y receptors / cellular response to purine-containing compound / G protein-coupled purinergic nucleotide receptor activity / positive regulation of inositol trisphosphate biosynthetic process / positive regulation of monoatomic ion transport / positive regulation of penile erection ...G protein-coupled ATP receptor activity / relaxation of muscle / P2Y receptors / cellular response to purine-containing compound / G protein-coupled purinergic nucleotide receptor activity / positive regulation of inositol trisphosphate biosynthetic process / positive regulation of monoatomic ion transport / positive regulation of penile erection ...G protein-coupled ATP receptor activity / relaxation of muscle /  A1 adenosine receptor binding / G protein-coupled ADP receptor activity / A1 adenosine receptor binding / G protein-coupled ADP receptor activity /  P2Y receptors / cellular response to purine-containing compound / G protein-coupled purinergic nucleotide receptor activity / positive regulation of inositol trisphosphate biosynthetic process / positive regulation of monoatomic ion transport / positive regulation of penile erection / negative regulation of norepinephrine secretion / glial cell migration / regulation of presynaptic cytosolic calcium ion concentration / G protein-coupled adenosine receptor signaling pathway / signal transduction involved in regulation of gene expression / positive regulation of hormone secretion / response to growth factor / cellular response to ATP / regulation of synaptic vesicle exocytosis / P2Y receptors / cellular response to purine-containing compound / G protein-coupled purinergic nucleotide receptor activity / positive regulation of inositol trisphosphate biosynthetic process / positive regulation of monoatomic ion transport / positive regulation of penile erection / negative regulation of norepinephrine secretion / glial cell migration / regulation of presynaptic cytosolic calcium ion concentration / G protein-coupled adenosine receptor signaling pathway / signal transduction involved in regulation of gene expression / positive regulation of hormone secretion / response to growth factor / cellular response to ATP / regulation of synaptic vesicle exocytosis /  eating behavior / monoatomic ion transport / response to mechanical stimulus / presynaptic active zone membrane / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / blood vessel diameter maintenance / establishment of localization in cell / protein localization to plasma membrane / eating behavior / monoatomic ion transport / response to mechanical stimulus / presynaptic active zone membrane / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / blood vessel diameter maintenance / establishment of localization in cell / protein localization to plasma membrane /  ADP binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / ADP binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits /  cilium / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / cilium / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion /  platelet activation / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / Inactivation, recovery and regulation of the phototransduction cascade / platelet activation / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / Inactivation, recovery and regulation of the phototransduction cascade /  heterotrimeric G-protein complex / G alpha (12/13) signalling events / heterotrimeric G-protein complex / G alpha (12/13) signalling events /  extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye /  signaling receptor activity / signaling receptor activity /  GTPase binding / Ca2+ pathway / GTPase binding / Ca2+ pathway /  cell body / phospholipase C-activating G protein-coupled receptor signaling pathway / regulation of cell shape / G alpha (i) signalling events / positive regulation of cytosolic calcium ion concentration / fibroblast proliferation / G alpha (s) signalling events / cell body / phospholipase C-activating G protein-coupled receptor signaling pathway / regulation of cell shape / G alpha (i) signalling events / positive regulation of cytosolic calcium ion concentration / fibroblast proliferation / G alpha (s) signalling events /  scaffold protein binding / G alpha (q) signalling events / scaffold protein binding / G alpha (q) signalling events /  postsynaptic membrane / basolateral plasma membrane / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / postsynaptic membrane / basolateral plasma membrane / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling /  postsynaptic density / cell surface receptor signaling pathway / positive regulation of ERK1 and ERK2 cascade / positive regulation of protein phosphorylation / apical plasma membrane / G protein-coupled receptor signaling pathway / protein heterodimerization activity / lysosomal membrane / postsynaptic density / cell surface receptor signaling pathway / positive regulation of ERK1 and ERK2 cascade / positive regulation of protein phosphorylation / apical plasma membrane / G protein-coupled receptor signaling pathway / protein heterodimerization activity / lysosomal membrane /  GTPase activity / GTPase activity /  dendrite / dendrite /  synapse / glutamatergic synapse / protein-containing complex binding / synapse / glutamatergic synapse / protein-containing complex binding /  cell surface cell surfaceSimilarity search - Function | ||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.9 Å cryo EM / Resolution: 2.9 Å | ||||||||||||||||||
 Authors Authors | Tan Q / Li B / Han S / Zhao Q / Wu B | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2023 Journal: Protein Cell / Year: 2023Title: Structural insights into signal transduction of the purinergic receptors P2Y1R and P2Y12R. Authors: Beibei Li / Shuo Han / Mu Wang / Yu Yu / Limin Ma / Xiaojing Chu / Qiuxiang Tan / Qiang Zhao / Beili Wu /  | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33503.map.gz emd_33503.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33503-v30.xml emd-33503-v30.xml emd-33503.xml emd-33503.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33503.png emd_33503.png | 43.9 KB | ||
| Others |  emd_33503_half_map_1.map.gz emd_33503_half_map_1.map.gz emd_33503_half_map_2.map.gz emd_33503_half_map_2.map.gz | 49.5 MB 49.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33503 http://ftp.pdbj.org/pub/emdb/structures/EMD-33503 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33503 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33503 | HTTPS FTP |
-Related structure data
| Related structure data |  7xxhMC  7xxiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33503.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33503.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
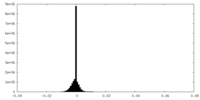
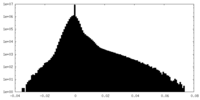
-Half map: #2
| File | emd_33503_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
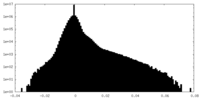
| Projections & Slices |
| ||||||||||||
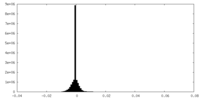
| Density Histograms |
-Half map: #1
| File | emd_33503_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The purinergic receptor P2Y1R in complex with 2MeSADP, G11 and scfv16
| Entire | Name: The purinergic receptor P2Y1R in complex with 2MeSADP, G11 and scfv16 |
|---|---|
| Components |
|
-Supramolecule #1: The purinergic receptor P2Y1R in complex with 2MeSADP, G11 and scfv16
| Supramolecule | Name: The purinergic receptor P2Y1R in complex with 2MeSADP, G11 and scfv16 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2-#5 / Details: scf16 was recombinantly expressed in sf9 cells |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10 kDa/nm |
-Macromolecule #1: P2Y purinoceptor 1
| Macromolecule | Name: P2Y purinoceptor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.296887 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: GAPTEVLWPA VPNGTDAAFL AGPGSSWGNS TVASTAAVSS SFKCALTKTG FQFYYLPAVY ILVFIIGFLG NSVAIWMFVF HMKPWSGIS VYMFNLALAD FLYVLTLPAL IFYYFNKTDW IFGDAMCKLQ RFIFHVNLYG SILFLTCISA HRYSGVVYPL K SLGRLKKK ...String: GAPTEVLWPA VPNGTDAAFL AGPGSSWGNS TVASTAAVSS SFKCALTKTG FQFYYLPAVY ILVFIIGFLG NSVAIWMFVF HMKPWSGIS VYMFNLALAD FLYVLTLPAL IFYYFNKTDW IFGDAMCKLQ RFIFHVNLYG SILFLTCISA HRYSGVVYPL K SLGRLKKK NAICISVLVW LIVVVAISPI LFYSGTGVRK NKTITCYDTT SDEYLRSYFI YSMCTTVAMF CVPLVLILGC YG LIVRALI YKDLDNSPLR RKSIYLVIIV LTVFAVSYIP FHVMKTMNLR ARLDFQTPAM CAFNDRVYAT YQVTRGLASL NSC VDPILY FLAGDTFRRR LSRATRKASR RSEANLQSKS EDMTLNILPE FKQNGDTSLE FLEVLFQGPG SWSHPQFEKG SGAG ASAGS WSHPQFEK UniProtKB: P2Y purinoceptor 1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(11) subunit alpha
| Macromolecule | Name: Guanine nucleotide-binding protein G(11) subunit alpha type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.385281 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLRRDKRDA RRELKLLLLG TGESGKSTFI KQMRIIHGAG YSEEDKRGFT KLVYQNIFTA MQAMIRAME TLKILYKYEQ NKANALLIRE VDVEKVTTFE HQYVSAIKTL WEDPGIQECY DRRREYQLSD SAKYYLTDVD R IATLGYLP ...String: MGCTLSAEDK AAVERSKMID RNLRRDKRDA RRELKLLLLG TGESGKSTFI KQMRIIHGAG YSEEDKRGFT KLVYQNIFTA MQAMIRAME TLKILYKYEQ NKANALLIRE VDVEKVTTFE HQYVSAIKTL WEDPGIQECY DRRREYQLSD SAKYYLTDVD R IATLGYLP TQQDVLRVRV PTTGIIEYPF DLENIIFRMV DVGGQRSERR KWIHCFENVT SIMFLVALSE YDQVLVESDN EN RMEESKA LFRTIITYPW FQNSSVILFL NKKDLLEDKI LYSHLVDYFP EFDGPQRDAQ AAREFILKMF VDLNPDSDKI IYS HFTCAT DTENIRFVFA AVKDTILQLN LKEYNLV |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.245805 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MHHHHHHSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSAS QDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT GYLSCCRFLD D NQIVTSSG ...String: MHHHHHHSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSAS QDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT GYLSCCRFLD D NQIVTSSG DTTCALWDIE TGQQTTTFTG HTGDVMSLSL APDTRLFVSG ACDASAKLWD VREGMCRQTF TGHESDINAI CF FPNGNAF ATGSDDATCR LFDLRADQEL MTYSHDNIIC GITSVSFSKS GRLLLAGYDD FNCNVWDALK ADRAGVLAGH DNR VSCLGV TDDGMAVATG SWDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scfv16
| Macromolecule | Name: scfv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.337307 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL |
-Macromolecule #6: 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate)
| Macromolecule | Name: 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate) type: ligand / ID: 6 / Number of copies: 1 / Formula: 6AD |
|---|---|
| Molecular weight | Theoretical: 473.293 Da |
| Chemical component information | 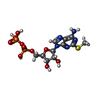 ChemComp-6AD: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Concentration: 2.0 mg/ml / Component - Formula: 150mM / Component - Name: sodium chloride |
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 1s. |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 2 / Number real images: 9856 / Average exposure time: 2.0 sec. / Average electron dose: 60.0 e/Å2 / Details: Images were collected in movie-mode |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 6184568 |
|---|---|
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: Details: 6OIJ for G protein |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final 3D classification | Software - Name: RELION (ver. 3.0) |
| Final angle assignment | Type: NOT APPLICABLE |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 659399 |
| Details | The selected images were 20eV using GIF-Quantum LS Imaging energy filter |
 Movie
Movie Controller
Controller

























 Z
Z Y
Y X
X