+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ADGRL3/Gi complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationlocomotion involved in locomotory behavior / maintenance of postsynaptic specialization structure / cell-cell adhesion via plasma-membrane adhesion molecules / positive regulation of synapse assembly / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / regulation of mitotic spindle organization ...locomotion involved in locomotory behavior / maintenance of postsynaptic specialization structure / cell-cell adhesion via plasma-membrane adhesion molecules / positive regulation of synapse assembly / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / regulation of mitotic spindle organization / cellular response to forskolin /  synapse assembly / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to cocaine / Regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / synapse organization / synapse assembly / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to cocaine / Regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / synapse organization /  neuron migration / Olfactory Signaling Pathway / G-protein beta/gamma-subunit complex binding / Activation of the phototransduction cascade / Schaffer collateral - CA1 synapse / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / response to peptide hormone / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / GDP binding / Inactivation, recovery and regulation of the phototransduction cascade / neuron migration / Olfactory Signaling Pathway / G-protein beta/gamma-subunit complex binding / Activation of the phototransduction cascade / Schaffer collateral - CA1 synapse / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / response to peptide hormone / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / GDP binding / Inactivation, recovery and regulation of the phototransduction cascade /  heterotrimeric G-protein complex / G alpha (12/13) signalling events / heterotrimeric G-protein complex / G alpha (12/13) signalling events /  extracellular vesicle / cell-cell junction / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / extracellular vesicle / cell-cell junction / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye /  GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway /  cell cortex / midbody / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / cell cortex / midbody / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events /  postsynaptic membrane / postsynaptic membrane /  carbohydrate binding / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / cell surface receptor signaling pathway / carbohydrate binding / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / cell surface receptor signaling pathway /  cell cycle / G protein-coupled receptor signaling pathway / lysosomal membrane / cell cycle / G protein-coupled receptor signaling pathway / lysosomal membrane /  axon / axon /  cell division / cell division /  GTPase activity / GTPase activity /  centrosome / centrosome /  synapse / glutamatergic synapse / synapse / glutamatergic synapse /  calcium ion binding / protein-containing complex binding / GTP binding / calcium ion binding / protein-containing complex binding / GTP binding /  nucleolus / magnesium ion binding / nucleolus / magnesium ion binding /  signal transduction / extracellular exosome / signal transduction / extracellular exosome /  nucleoplasm / nucleoplasm /  membrane / membrane /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
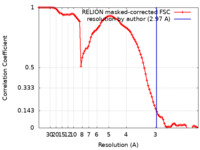
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.97 Å cryo EM / Resolution: 2.97 Å | |||||||||
 Authors Authors | He Y / Qian Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structural insights into adhesion GPCR ADGRL3 activation and G, G, G, and G coupling. Authors: Yu Qian / Zhengxiong Ma / Chunhong Liu / Xinzhi Li / Xinyan Zhu / Na Wang / Zhenmei Xu / Ruixue Xia / Jiale Liang / Yaning Duan / Han Yin / Yangjie Xiong / Anqi Zhang / Changyou Guo / Zheng ...Authors: Yu Qian / Zhengxiong Ma / Chunhong Liu / Xinzhi Li / Xinyan Zhu / Na Wang / Zhenmei Xu / Ruixue Xia / Jiale Liang / Yaning Duan / Han Yin / Yangjie Xiong / Anqi Zhang / Changyou Guo / Zheng Chen / Zhiwei Huang / Yuanzheng He /  Abstract: Adhesion G-protein-coupled receptors (aGPCRs) play key roles in a diversity of physiologies. A hallmark of aGPCR activation is the removal of the inhibitory GAIN domain and the dipping of the cleaved ...Adhesion G-protein-coupled receptors (aGPCRs) play key roles in a diversity of physiologies. A hallmark of aGPCR activation is the removal of the inhibitory GAIN domain and the dipping of the cleaved stalk peptide into the ligand-binding pocket of receptors; however, the detailed mechanism remains obscure. Here, we present cryoelectron microscopy (cryo-EM) structures of ADGRL3 in complex with G, G, G, and G. The structures reveal unique ligand-engaging mode, distinctive activation conformation, and key mechanisms of aGPCR activation. The structures also reveal the uncharted structural information of GPCR/G coupling. A comparison of G, G, G, and G engagements with ADGRL3 reveals the key determinant of G-protein coupling on the far end of αH5 of Gα. A detailed analysis of the engagements allows us to design mutations that specifically enhance one pathway over others. Taken together, our study lays the groundwork for understanding aGPCR activation and G-protein-coupling selectivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32890.map.gz emd_32890.map.gz | 56.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32890-v30.xml emd-32890-v30.xml emd-32890.xml emd-32890.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32890_fsc.xml emd_32890_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_32890.png emd_32890.png | 73.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32890 http://ftp.pdbj.org/pub/emdb/structures/EMD-32890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32890 | HTTPS FTP |
-Related structure data
| Related structure data |  7wybMC  7wy5C  7wy8C  7x10C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32890.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32890.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : GPCR/G-protein complex
| Entire | Name: GPCR/G-protein complex |
|---|---|
| Components |
|
-Supramolecule #1: GPCR/G-protein complex
| Supramolecule | Name: GPCR/G-protein complex / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: GPCR/G-protein complex
| Supramolecule | Name: GPCR/G-protein complex / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: ADGRL3
| Supramolecule | Name: ADGRL3 / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #5 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.414047 KDa |
| Recombinant expression | Organism:  Insect BA phytoplasma (bacteria) Insect BA phytoplasma (bacteria) |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.915496 KDa |
| Recombinant expression | Organism:  Insect BA phytoplasma (bacteria) Insect BA phytoplasma (bacteria) |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Insect BA phytoplasma (bacteria) Insect BA phytoplasma (bacteria) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.277299 KDa |
| Recombinant expression | Organism:  Insect BA phytoplasma (bacteria) Insect BA phytoplasma (bacteria) |
| Sequence | String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL |
-Macromolecule #5: Isoform 3 of Adhesion G protein-coupled receptor L3
| Macromolecule | Name: Isoform 3 of Adhesion G protein-coupled receptor L3 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 172.044625 KDa |
| Recombinant expression | Organism:  Insect BA phytoplasma (bacteria) Insect BA phytoplasma (bacteria) |
| Sequence | String: MWPPQLLILT MLLAPVVHGG KHNERHPALA APLRHAERSP GGALPPRHLL QQPAAERSTA HRGQGPRGAA RGVRGPGAPG AQIAAQAFS RAPIPMAVVR RELSCESYPI ELRCPGTDVI MIESANYGRT DDKICDSDPA QMENIRCYLP DAYKIMSQRC N NRTQCAVV ...String: MWPPQLLILT MLLAPVVHGG KHNERHPALA APLRHAERSP GGALPPRHLL QQPAAERSTA HRGQGPRGAA RGVRGPGAPG AQIAAQAFS RAPIPMAVVR RELSCESYPI ELRCPGTDVI MIESANYGRT DDKICDSDPA QMENIRCYLP DAYKIMSQRC N NRTQCAVV AGPDVFPDPC PGTYKYLEVQ YECVPYKVEQ KVFLCPGLLK GVYQSEHLFE SDHQSGAWCK DPLQASDKIY YM PWTPYRT DTLTEYSSKD DFIAGRPTTT YKLPHRVDGT GFVVYDGALF FNKERTRNIV KFDLRTRIKS GEAIIANANY HDT SPYRWG GKSDIDLAVD ENGLWVIYAT EQNNGKIVIS QLNPYTLRIE GTWDTAYDKR SASNAFMICG ILYVVKSVYE DDDN EATGN KIDYIYNTDQ SKDSLVDVPF PNSYQYIAAV DYNPRDNLLY VWNNYHVVKY SLDFGPLDSR SGPVHHGQVS YISPP IHLD SELERPPVRG ISTTGSLGMG STTTSTTLRT TTWNIGRSTT ASLPGRRNRS TSTPSPAVEV LDDVTTHLPS AASQIP AME ESCEAVEARE IMWFKTRQGQ VAKQPCPAGT IGVSTYLCLA PDGIWDPQGP DLSNCSSPWV NHITQKLKSG ETAANIA RE LAEQTRNHLN AGDITYSVRA MDQLVGLLDV QLRNLTPGGK DSAARSLNKL QKRERSCRAY VQAMVETVNN LLQPQALN A WRDLTTSDQL RAATMLLDTV EESAFVLADN LLKTDIVREN TDNIQLEVAR LSTEGNLEDL KFPENMGHGS TIQLSANTL KQNGRNGEIR VAFVLYNNLG PYLSTENASM KLGTEAMSTN HSVIVNSPVI TAAINKEFSN KVYLADPVVF TVKHIKQSEE NFNPNCSFW SYSKRTMTGY WSTQGCRLLT TNKTHTTCSC NHLTNFAVLM AHVEVKHSDA VHDLLLDVIT WVGILLSLVC L LICIFTFC FFRGLQSDRN TIHKNLCISL FVAELLFLIG INRTDQPIAC AVFAALLHFF FLAAFTWMFL EGVQLYIMLV EV FESEHSR RKYFYLVGYG MPALIVAVSA AVDYRSYGTD KVCWLRLDTY FIWSFIGPAT LIIMLNVIFL GIALYKMFHH TAI LKPESG CLDNINYEDN RPFIKSWVIG AIALLCLLGL TWAFGLMYIN ESTVIMAYLF TIFNSLQGMF IFIFHCVLQK KVRK EYGKC LRTHCCSGKS TESSIGSGKT SGSRTPGRYS TGSQSRIRRM WNDTVRKQSE SSFITGDINS SASLNREPYR ETKGL LNNA RDTSVMDTLP LNGNHGNSYS IAGGEYLSNC VQIIDRGYNH NETALEKKIL KELTSNYIPS YLNNHERSSE QNRNMM NKL VNNLGSGSED DAIVLDDAAS FNHEESLGLE LIHEESDAPL LPPRVYSTDN HQPHHYSRRR FPQDHSESFF PLLTDEH TE DLQSPHRDSL YTSMPALAGV PAADSVTTST QTEAAAAKGG DAEDVYYKSM PNLGSRNHVH PLHAYYQLGR GSSDGFIV P PNKDGASPEG TSKGPAHLVT SL |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller