[English] 日本語
 Yorodumi
Yorodumi- EMDB-3280: Biochemical and structural characterization of the yeast Sdo1p su... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3280 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Biochemical and structural characterization of the yeast Sdo1p suggests a surveillance role in the 60S ribosomal subunit maturation | |||||||||
 Map data Map data | Reconstruction of 60S-Sdo1p complex. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Ribosome biogenesis / Ribosome biogenesis /  SBDS / SDS / Sdo1 / cryo-electron microscopy (cryo-EM) SBDS / SDS / Sdo1 / cryo-electron microscopy (cryo-EM) | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
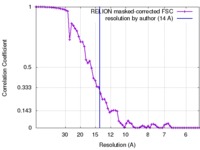
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 14.0 Å cryo EM / Resolution: 14.0 Å | |||||||||
 Authors Authors | Ma CY / Yan KG / Tan D / Li NN / Zhang YX / Yuan Y / Li ZF / Dong MQ / Lei JL / Gao N | |||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2016 Journal: Protein Cell / Year: 2016Title: Structural dynamics of the yeast Shwachman-Diamond syndrome protein (Sdo1) on the ribosome and its implication in the 60S subunit maturation. Authors: Chengying Ma / Kaige Yan / Dan Tan / Ningning Li / Yixiao Zhang / Yi Yuan / Zhifei Li / Meng-Qiu Dong / Jianlin Lei / Ning Gao /  Abstract: The human Shwachman-Diamond syndrome (SDS) is an autosomal recessive disease caused by mutations in a highly conserved ribosome assembly factor SBDS. The functional role of SBDS is to cooperate with ...The human Shwachman-Diamond syndrome (SDS) is an autosomal recessive disease caused by mutations in a highly conserved ribosome assembly factor SBDS. The functional role of SBDS is to cooperate with another assembly factor, elongation factor 1-like (Efl1), to promote the release of eukaryotic initiation factor 6 (eIF6) from the late-stage cytoplasmic 60S precursors. In the present work, we characterized, both biochemically and structurally, the interaction between the 60S subunit and SBDS protein (Sdo1p) from yeast. Our data show that Sdo1p interacts tightly with the mature 60S subunit in vitro through its domain I and II, and is capable of bridging two 60S subunits to form a stable 2:2 dimer. Structural analysis indicates that Sdo1p bind to the ribosomal P-site, in the proximity of uL16 and uL5, and with direct contact to H69 and H38. The dynamic nature of Sdo1p on the 60S subunit, together with its strategic binding position, suggests a surveillance role of Sdo1p in monitoring the conformational maturation of the ribosomal P-site. Altogether, our data support a conformational signal-relay cascade during late-stage 60S maturation, involving uL16, Sdo1p, and Efl1p, which interrogates the functional P-site to control the departure of the anti-association factor eIF6. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3280.map.gz emd_3280.map.gz | 16.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3280-v30.xml emd-3280-v30.xml emd-3280.xml emd-3280.xml | 8.7 KB 8.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3280_fsc.xml emd_3280_fsc.xml | 6.3 KB | Display |  FSC data file FSC data file |
| Images |  EMD-3280_60S-Sdo1p.jpg EMD-3280_60S-Sdo1p.jpg emd_3280.png emd_3280.png | 115.8 KB 104.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3280 http://ftp.pdbj.org/pub/emdb/structures/EMD-3280 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3280 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3280 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3280.map.gz / Format: CCP4 / Size: 21.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3280.map.gz / Format: CCP4 / Size: 21.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of 60S-Sdo1p complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.76 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The dimeric structure of the yeast 60S ribosomal subunits bound w...
| Entire | Name: The dimeric structure of the yeast 60S ribosomal subunits bound with Sdo1p |
|---|---|
| Components |
|
-Supramolecule #1000: The dimeric structure of the yeast 60S ribosomal subunits bound w...
| Supramolecule | Name: The dimeric structure of the yeast 60S ribosomal subunits bound with Sdo1p type: sample / ID: 1000 / Oligomeric state: two Sdo1p binding to two 60S subunit / Number unique components: 2 |
|---|
-Supramolecule #1: Sdo1p bound 60S subunit
| Supramolecule | Name: Sdo1p bound 60S subunit / type: complex / ID: 1 / Recombinant expression: No / Ribosome-details: ribosome-eukaryote: LSU 60S |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) / synonym: yeast Saccharomyces cerevisiae (brewer's yeast) / synonym: yeast |
-Macromolecule #1: Sdo1p
| Macromolecule | Name: Sdo1p / type: protein_or_peptide / ID: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) / synonym: yeast Saccharomyces cerevisiae (brewer's yeast) / synonym: yeast |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm Bright-field microscopy / Cs: 2.0 mm |
| Sample stage | Specimen holder: Gatan CT3500 / Specimen holder model: OTHER |
| Date | Dec 8, 2011 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (4k x 4k) |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID:  3o58 |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller