+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Porcine epidemic diarrhea virus core polymerase complex | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  Coronavirus / nsp12 / Coronavirus / nsp12 /  RNA polymerase / RNA polymerase /  PEDV / PEDV /  REPLICATION REPLICATION | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell membrane /  endoplasmic reticulum-Golgi intermediate compartment / viral genome replication / 3'-5'-RNA exonuclease activity / host cell endoplasmic reticulum-Golgi intermediate compartment / endoplasmic reticulum-Golgi intermediate compartment / viral genome replication / 3'-5'-RNA exonuclease activity / host cell endoplasmic reticulum-Golgi intermediate compartment /  omega peptidase activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / symbiont-mediated perturbation of host ubiquitin-like protein modification / omega peptidase activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / symbiont-mediated perturbation of host ubiquitin-like protein modification /  endonuclease activity / endonuclease activity /  mRNA (nucleoside-2'-O-)-methyltransferase activity ...host cell membrane / mRNA (nucleoside-2'-O-)-methyltransferase activity ...host cell membrane /  endoplasmic reticulum-Golgi intermediate compartment / viral genome replication / 3'-5'-RNA exonuclease activity / host cell endoplasmic reticulum-Golgi intermediate compartment / endoplasmic reticulum-Golgi intermediate compartment / viral genome replication / 3'-5'-RNA exonuclease activity / host cell endoplasmic reticulum-Golgi intermediate compartment /  omega peptidase activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / symbiont-mediated perturbation of host ubiquitin-like protein modification / omega peptidase activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / symbiont-mediated perturbation of host ubiquitin-like protein modification /  endonuclease activity / endonuclease activity /  mRNA (nucleoside-2'-O-)-methyltransferase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / cysteine-type deubiquitinase activity / mRNA (nucleoside-2'-O-)-methyltransferase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / cysteine-type deubiquitinase activity /  RNA helicase activity / host cell perinuclear region of cytoplasm / RNA helicase activity / host cell perinuclear region of cytoplasm /  viral protein processing / viral protein processing /  lyase activity / induction by virus of host autophagy / cysteine-type endopeptidase activity / lyase activity / induction by virus of host autophagy / cysteine-type endopeptidase activity /  RNA-dependent RNA polymerase activity / DNA-templated transcription / RNA-dependent RNA polymerase activity / DNA-templated transcription /  proteolysis / proteolysis /  RNA binding / zinc ion binding / RNA binding / zinc ion binding /  ATP binding / ATP binding /  membrane / identical protein binding membrane / identical protein bindingSimilarity search - Function | ||||||||||||
| Biological species |   Porcine epidemic diarrhea virus / synthetic construct (others) Porcine epidemic diarrhea virus / synthetic construct (others) | ||||||||||||
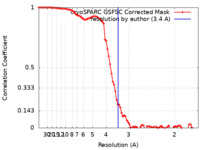
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.3 Å cryo EM / Resolution: 3.3 Å | ||||||||||||
 Authors Authors | Anderson TK / Kirchdoerfer RN | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: An alphacoronavirus polymerase structure reveals conserved co-factor functions. Authors: Thomas K Anderson / Peter J Hoferle / Kenneth W Lee / Joshua J Coon / Robert N Kirchdoerfer /  Abstract: Coronaviruses are a diverse subfamily of viruses containing pathogens of humans and animals. This subfamily of viruses replicates their RNA genomes using a core polymerase complex composed of viral ...Coronaviruses are a diverse subfamily of viruses containing pathogens of humans and animals. This subfamily of viruses replicates their RNA genomes using a core polymerase complex composed of viral non-structural proteins: nsp7, nsp8 and nsp12. Most of our understanding of coronavirus molecular biology comes from the betacoronaviruses like SARS-CoV and SARS-CoV-2, the latter of which is the causative agent of COVID-19. In contrast, members of the alphacoronavirus genus are relatively understudied despite their importance in human and animal health. Here we have used cryo-electron microscopy to determine the structure of the alphacoronavirus porcine epidemic diarrhea virus (PEDV) core polymerase complex bound to RNA. Our structure shows an unexpected nsp8 stoichiometry in comparison to other published coronavirus polymerase structures. Biochemical analysis shows that the N-terminal extension of one nsp8 is not required for RNA synthesis for alpha and betacoronaviruses as previously hypothesized. Our work shows the importance of studying diverse coronaviruses to reveal aspects of coronavirus replication while also identifying areas of conservation to be targeted by antiviral drugs. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29779.map.gz emd_29779.map.gz | 32.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29779-v30.xml emd-29779-v30.xml emd-29779.xml emd-29779.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29779_fsc.xml emd_29779_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_29779.png emd_29779.png | 63 KB | ||
| Masks |  emd_29779_msk_1.map emd_29779_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29779.cif.gz emd-29779.cif.gz | 7 KB | ||
| Others |  emd_29779_half_map_1.map.gz emd_29779_half_map_1.map.gz emd_29779_half_map_2.map.gz emd_29779_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29779 http://ftp.pdbj.org/pub/emdb/structures/EMD-29779 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29779 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29779 | HTTPS FTP |
-Related structure data
| Related structure data |  8g6rMC  8urbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29779.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29779.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.834 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29779_msk_1.map emd_29779_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29779_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29779_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Core replication complex of porcine epidemic diarrhea virus compo...
| Entire | Name: Core replication complex of porcine epidemic diarrhea virus composed of nsp12, nsp7, nsp8, and a short RNA substrate |
|---|---|
| Components |
|
-Supramolecule #1: Core replication complex of porcine epidemic diarrhea virus compo...
| Supramolecule | Name: Core replication complex of porcine epidemic diarrhea virus composed of nsp12, nsp7, nsp8, and a short RNA substrate type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Porcine epidemic diarrhea virus Porcine epidemic diarrhea virus |
| Molecular weight | Theoretical: 185 KDa |
-Macromolecule #1: nsp12
| Macromolecule | Name: nsp12 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Porcine epidemic diarrhea virus Porcine epidemic diarrhea virus |
| Molecular weight | Theoretical: 104.939398 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: DMAYLNRVRG SSAARLEPCN GTDTQHVYRA FDIYNKDVAC LGKFLKVNCV RLKNLDKHDA FYVVKRCTKS AMEHEQSIYS RLEKCGAVA EHDFFTWKDG RAIYGNVCRK DLTEYTMMDL CYALRNFDEN NCDVLKSILI KVGACEESYF NNKVWFDPVE N EDIHRVYA ...String: DMAYLNRVRG SSAARLEPCN GTDTQHVYRA FDIYNKDVAC LGKFLKVNCV RLKNLDKHDA FYVVKRCTKS AMEHEQSIYS RLEKCGAVA EHDFFTWKDG RAIYGNVCRK DLTEYTMMDL CYALRNFDEN NCDVLKSILI KVGACEESYF NNKVWFDPVE N EDIHRVYA LLGTIVSRAM LKCVKFCDAM VEQGIVGVVT LDNQDLNGDF YDFGDFTCSI KGMGIPICTS YYSYMMPVMG MT NCLASEC FVKSDIFGED FKSYDLLEYD FTEHKTALFN KYFKYWGLQY HPNCVDCSDE QCIVHCANFN TLFSTTIPIT AFG PLCRKC WIDGVPLVTT AGYHFKQLGI VWNNDLNLHS SRLSINELLQ FCSDPALLIA SSPALVDQRT VCFSVAALGT GMTN QTVKP GHFNKEFYDF LLEQGFFSEG SELTLKHFFF AQKGDAAVKD FDYYRYNRPT VLDICQARVV YQIVQRYFDI YEGGC ITAK EVVVTNLNKS AGYPLNKFGK AGLYYESLSY EEQDELYAYT KRNILPTMTQ LNLKYAISGK ERARTVGGVS LLSTMT TRQ YHQKHLKSIV NTRGASVVIG TTKFYGGWDN MLKNLIDGVE NPCLMGWDYP KCDRALPNMI RMISAMILGS KHTTCCS ST DRFFRLCNEL AQVLTEVVYS NGGFYLKPGG TTSGDATTAY ANSVFNIFQA VSANVNKLLS VDSNVCHNLE VKQLQRKL Y ECCYRSTTVD DQFVVEYYGY LRKHFSMMIL SDDGVVCYNN DYASLGYVAD LNAFKAVLYY QNNVFMSASK CWIEPDINK GPHEFCSQHT MQIVDKDGTY YLPYPDPSRI LSAGVFVDDV VKTDAVVLLE RYVSLAIDAY PLSKHENPEY KKVFYVLLDW VKHLYKTLN AGVLESFSVT LLEDSTAKFW DESFYANMYE KS UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #2: nsp8
| Macromolecule | Name: nsp8 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Porcine epidemic diarrhea virus Porcine epidemic diarrhea virus |
| Molecular weight | Theoretical: 12.561573 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: RKSKVVSAMH SLLFGMLRRL DMSSVDTILN LAKDGVVPLS VIPAVSATKL NIVTSDIDSY NRIQREGCVH YAGTIWNIID IKDNDGKVV HVKEVTAQNA ESLSWPLVLG CERIV UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #3: nsp7
| Macromolecule | Name: nsp7 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Porcine epidemic diarrhea virus Porcine epidemic diarrhea virus |
| Molecular weight | Theoretical: 6.88201 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: KLTDIKCSNV VLLGCLSSMN VSANSTEWAY CVDLHNKINL CNDPEKAQEM LLALLAFFLS KN UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #4: RNA (5'-R(P*AP*AP*GP*AP*AP*GP*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP...
| Macromolecule | Name: RNA (5'-R(P*AP*AP*GP*AP*AP*GP*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*A)-3') type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 6.406927 KDa |
| Sequence | String: AAGAAGCUAU UAAAAUCACA |
-Macromolecule #5: RNA (5'-R(P*GP*GP*UP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP...
| Macromolecule | Name: RNA (5'-R(P*GP*GP*UP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*U)-3') type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 6.364735 KDa |
| Sequence | String: GGUUGUGAUU UUAAUAGCUU |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 1.2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X