[English] 日本語
 Yorodumi
Yorodumi- EMDB-2975: Cryo electron microscopy of yeast vacuolar ATPase proton channel ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2975 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo electron microscopy of yeast vacuolar ATPase proton channel sector | |||||||||
 Map data Map data | Reconstruction of yeast V-ATPase membrane sector (Vo) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  yeast vacuolar ATPase / proton channel sector / reversible disassembly / yeast vacuolar ATPase / proton channel sector / reversible disassembly /  membrane protein membrane protein | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
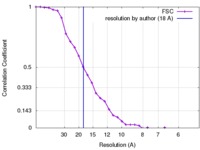
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 18.0 Å cryo EM / Resolution: 18.0 Å | |||||||||
 Authors Authors | Couoh-Cardel S / Milgrom E / Wilkens S | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2015 Journal: J Biol Chem / Year: 2015Title: Affinity Purification and Structural Features of the Yeast Vacuolar ATPase Vo Membrane Sector. Authors: Sergio Couoh-Cardel / Elena Milgrom / Stephan Wilkens /  Abstract: The membrane sector (Vo) of the proton pumping vacuolar ATPase (V-ATPase, V1Vo-ATPase) from Saccharomyces cerevisiae was purified to homogeneity, and its structure was characterized by EM of single ...The membrane sector (Vo) of the proton pumping vacuolar ATPase (V-ATPase, V1Vo-ATPase) from Saccharomyces cerevisiae was purified to homogeneity, and its structure was characterized by EM of single molecules and two-dimensional crystals. Projection images of negatively stained Vo two-dimensional crystals showed a ring-like structure with a large asymmetric mass at the periphery of the ring. A cryo-EM reconstruction of Vo from single-particle images showed subunits a and d in close contact on the cytoplasmic side of the proton channel. A comparison of three-dimensional reconstructions of free Vo and Vo as part of holo V1Vo revealed that the cytoplasmic N-terminal domain of subunit a (aNT) must undergo a large conformational change upon enzyme disassembly or (re)assembly from Vo, V1, and subunit C. Isothermal titration calorimetry using recombinant subunit d and aNT revealed that the two proteins bind each other with a Kd of ~5 μm. Treatment of the purified Vo sector with 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-rac-(1-glycerol)] resulted in selective release of subunit d, allowing purification of a VoΔd complex. Passive proton translocation assays revealed that both Vo and VoΔd are impermeable to protons. We speculate that the structural change in subunit a upon release of V1 from Vo during reversible enzyme dissociation plays a role in blocking passive proton translocation across free Vo and that the interaction between aNT and d seen in free Vo functions to stabilize the Vo sector for efficient reassembly of V1Vo. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2975.map.gz emd_2975.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2975-v30.xml emd-2975-v30.xml emd-2975.xml emd-2975.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_2975_fsc.xml emd_2975_fsc.xml | 3.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_2975.tif emd_2975.tif | 86.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2975 http://ftp.pdbj.org/pub/emdb/structures/EMD-2975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2975 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2975.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2975.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of yeast V-ATPase membrane sector (Vo) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Membrane sector (Vo) of yeast vacuolar ATPase
| Entire | Name: Membrane sector (Vo) of yeast vacuolar ATPase |
|---|---|
| Components |
|
-Supramolecule #1000: Membrane sector (Vo) of yeast vacuolar ATPase
| Supramolecule | Name: Membrane sector (Vo) of yeast vacuolar ATPase / type: sample / ID: 1000 / Details: Sample was monodisperse / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 530 KDa / Theoretical: 320 KDa / Method: Small Angle X-ray Scattering |
-Macromolecule #1: Vacuolar ATPase membrane sector
| Macromolecule | Name: Vacuolar ATPase membrane sector / type: protein_or_peptide / ID: 1 / Name.synonym: Vo Details: V-ATPase membrane sector composed of subunits a, c, c', c'', d, e in the ratio 1:8:1:1:1:1 Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) / synonym: Baker's yeast / Location in cell: vacuole Saccharomyces cerevisiae (brewer's yeast) / synonym: Baker's yeast / Location in cell: vacuole |
| Molecular weight | Experimental: 530 KDa / Theoretical: 320 KDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 10 mM TRIS-HCl, 10 mM BME, 0.5 mM EGTA, 0.1 % dodecyl-beta-D-maltoside |
| Grid | Details: 400 mesh holey carbon (C-flat 2/2) glow discharged in air |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 90 K / Instrument: HOMEMADE PLUNGER / Method: 5 sec blot in cold room |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 57200 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 40000 Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Min: 97 K / Max: 113 K / Average: 103 K |
| Alignment procedure | Legacy - Astigmatism: Astigmatism was corrected at 200,000 times magnification over carbon film using live power spectrum |
| Date | Nov 10, 2010 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Digitization - Sampling interval: 15 µm / Average electron dose: 15 e/Å2 / Bits/pixel: 8 |
 Movie
Movie Controller
Controller