+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of yeast recombination mediator Rad52 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Recombination mediator protein /  DNA repair / Apo structure / DNA repair / Apo structure /  Decamer / Decamer /  RECOMBINATION RECOMBINATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationHDR through Single Strand Annealing (SSA) / meiotic joint molecule formation / double-strand break repair via single-strand annealing /  DNA amplification / DNA/DNA annealing activity / DNA recombinase assembly / SUMOylation of DNA damage response and repair proteins / DNA amplification / DNA/DNA annealing activity / DNA recombinase assembly / SUMOylation of DNA damage response and repair proteins /  mitotic recombination / DNA strand exchange activity / telomere maintenance via recombination ...HDR through Single Strand Annealing (SSA) / meiotic joint molecule formation / double-strand break repair via single-strand annealing / mitotic recombination / DNA strand exchange activity / telomere maintenance via recombination ...HDR through Single Strand Annealing (SSA) / meiotic joint molecule formation / double-strand break repair via single-strand annealing /  DNA amplification / DNA/DNA annealing activity / DNA recombinase assembly / SUMOylation of DNA damage response and repair proteins / DNA amplification / DNA/DNA annealing activity / DNA recombinase assembly / SUMOylation of DNA damage response and repair proteins /  mitotic recombination / DNA strand exchange activity / telomere maintenance via recombination / double-strand break repair via break-induced replication / mitotic recombination / DNA strand exchange activity / telomere maintenance via recombination / double-strand break repair via break-induced replication /  postreplication repair / mitochondrial DNA repair / nuclear chromosome / double-strand break repair via homologous recombination / postreplication repair / mitochondrial DNA repair / nuclear chromosome / double-strand break repair via homologous recombination /  mitochondrion / mitochondrion /  nucleus nucleusSimilarity search - Function | |||||||||
| Biological species |   Saccharomyces cerevisiae S288C (yeast) Saccharomyces cerevisiae S288C (yeast) | |||||||||
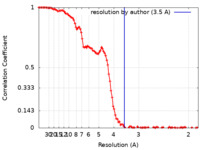
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Deveryshetty J / Basore K / Rau M / Fitzpatrick JAJ / Antony E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Yeast Rad52 is a homodecamer and possesses BRCA2-like bipartite Rad51 binding modes. Authors: Jaigeeth Deveryshetty / Rahul Chadda / Jenna R Mattice / Simrithaa Karunakaran / Michael J Rau / Katherine Basore / Nilisha Pokhrel / Noah Englander / James A J Fitzpatrick / Brian Bothner / Edwin Antony /  Abstract: Homologous recombination (HR) is an essential double-stranded DNA break repair pathway. In HR, Rad52 facilitates the formation of Rad51 nucleoprotein filaments on RPA-coated ssDNA. Here, we decipher ...Homologous recombination (HR) is an essential double-stranded DNA break repair pathway. In HR, Rad52 facilitates the formation of Rad51 nucleoprotein filaments on RPA-coated ssDNA. Here, we decipher how Rad52 functions using single-particle cryo-electron microscopy and biophysical approaches. We report that Rad52 is a homodecameric ring and each subunit possesses an ordered N-terminal and disordered C-terminal half. An intrinsic structural asymmetry is observed where a few of the C-terminal halves interact with the ordered ring. We describe two conserved charged patches in the C-terminal half that harbor Rad51 and RPA interacting motifs. Interactions between these patches regulate ssDNA binding. Surprisingly, Rad51 interacts with Rad52 at two different bindings sites: one within the positive patch in the disordered C-terminus and the other in the ordered ring. We propose that these features drive Rad51 nucleation onto a single position on the DNA to promote formation of uniform pre-synaptic Rad51 filaments in HR. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29695.map.gz emd_29695.map.gz | 96.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29695-v30.xml emd-29695-v30.xml emd-29695.xml emd-29695.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29695_fsc.xml emd_29695_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_29695.png emd_29695.png | 65.3 KB | ||
| Filedesc metadata |  emd-29695.cif.gz emd-29695.cif.gz | 6.4 KB | ||
| Others |  emd_29695_half_map_1.map.gz emd_29695_half_map_1.map.gz emd_29695_half_map_2.map.gz emd_29695_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29695 http://ftp.pdbj.org/pub/emdb/structures/EMD-29695 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29695 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29695 | HTTPS FTP |
-Related structure data
| Related structure data |  8g3gMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29695.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29695.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.94 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29695_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
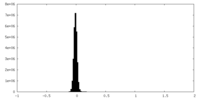
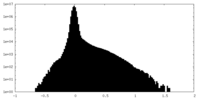
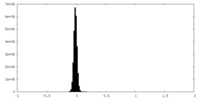
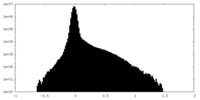
| Density Histograms |
-Half map: #1
| File | emd_29695_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rad52 decamer
| Entire | Name: Rad52 decamer |
|---|---|
| Components |
|
-Supramolecule #1: Rad52 decamer
| Supramolecule | Name: Rad52 decamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Full length yeast Rad52 expressed with chitin binding domain (CBD) tag. CBD was removed by thiol induced cleavage by intein. |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae S288C (yeast) / Location in cell: Nucleus Saccharomyces cerevisiae S288C (yeast) / Location in cell: Nucleus |
| Molecular weight | Theoretical: 520 kDa/nm |
-Macromolecule #1: DNA repair and recombination protein RAD52
| Macromolecule | Name: DNA repair and recombination protein RAD52 / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae S288C (yeast) Saccharomyces cerevisiae S288C (yeast) |
| Molecular weight | Theoretical: 52.476496 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MNEIMDMDEK KPVFGNHSED IQTKLDKKLG PEYISKRVGF GTSRIAYIEG WRVINLANQI FGYNGWSTEV KSVVIDFLDE RQGKFSIGC TAIVRVTLTS GTYREDIGYG TVENERRKPA AFERAKKSAV TDALKRSLRG FGNALGNCLY DKDFLAKIDK V KFDPPDFD ...String: MNEIMDMDEK KPVFGNHSED IQTKLDKKLG PEYISKRVGF GTSRIAYIEG WRVINLANQI FGYNGWSTEV KSVVIDFLDE RQGKFSIGC TAIVRVTLTS GTYREDIGYG TVENERRKPA AFERAKKSAV TDALKRSLRG FGNALGNCLY DKDFLAKIDK V KFDPPDFD ENNLFRPTDE ISESSRTNTL HENQEQQQYP NKRRQLTKVT NTNPDSTKNL VKIENTVSRG TPMMAAPAEA NS KNSSNKD TDLKSLDASK QDQDDLLDDS LMFSDDFQDD DLINMGNTNS NVLTTEKDPV VAKQSPTASS NPEAEQITFV TAK AATSVQ NERYIGEESI FDPKYQAQSI RHTVDQTTSK HIPASVLKDK TMTTARDSVY EKFAPKGKQL SMKNNDKELG PHML EGAGN QVPRETTPIK TNATAFPPAA APRFAPPSKV VHPNGNGAVP AVPQQRSTRR EVGRPKINPL HARKPT UniProtKB: DNA repair and recombination protein RAD52 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 97 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 150000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 150000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.7 e/Å2 |
- Image processing
Image processing
-Atomic model buiding 1
| Details | Initial model was built de novo in ModelAngelo tool. the best-looking subunit was real space refined in Phenix, followed by manual building in Coot. Later C10 symmetry was applied, and real space refined in Phenix. |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-8g3g: |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X