[English] 日本語
 Yorodumi
Yorodumi- EMDB-28793: Voltage-gated potassium channel Kv3.1 with novel positive modulat... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Voltage-gated potassium channel Kv3.1 with novel positive modulator (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one (compound 4) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  ion channel / positive modulator / voltage gated / ion channel / positive modulator / voltage gated /  voltage gated potassium channel / voltage gated potassium channel /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to nerve growth factor / globus pallidus development / response to light intensity / response to potassium ion / response to auditory stimulus / response to fibroblast growth factor / corpus callosum development / voltage-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / positive regulation of potassium ion transmembrane transport / Voltage gated Potassium channels ...response to nerve growth factor / globus pallidus development / response to light intensity / response to potassium ion / response to auditory stimulus / response to fibroblast growth factor / corpus callosum development / voltage-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / positive regulation of potassium ion transmembrane transport / Voltage gated Potassium channels / delayed rectifier potassium channel activity / optic nerve development / neuronal cell body membrane /  voltage-gated potassium channel activity / response to amine / voltage-gated potassium channel activity / response to amine /  kinesin binding / kinesin binding /  calyx of Held / calyx of Held /  axolemma / axon terminus / potassium ion transmembrane transport / axolemma / axon terminus / potassium ion transmembrane transport /  voltage-gated potassium channel complex / dendrite membrane / cerebellum development / protein tetramerization / protein homooligomerization / potassium ion transport / response to toxic substance / cellular response to xenobiotic stimulus / voltage-gated potassium channel complex / dendrite membrane / cerebellum development / protein tetramerization / protein homooligomerization / potassium ion transport / response to toxic substance / cellular response to xenobiotic stimulus /  presynaptic membrane / presynaptic membrane /  postsynaptic membrane / transmembrane transporter binding / postsynaptic membrane / transmembrane transporter binding /  cell surface / cell surface /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.92 Å cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Chen Y / Ishchenko A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Identification, structural, and biophysical characterization of a positive modulator of human Kv3.1 channels. Authors: Yun-Ting Chen / Mee Ra Hong / Xin-Jun Zhang / James Kostas / Yuxing Li / Richard L Kraus / Vincent P Santarelli / Deping Wang / Yacob Gomez-Llorente / Alexei Brooun / Corey Strickland / ...Authors: Yun-Ting Chen / Mee Ra Hong / Xin-Jun Zhang / James Kostas / Yuxing Li / Richard L Kraus / Vincent P Santarelli / Deping Wang / Yacob Gomez-Llorente / Alexei Brooun / Corey Strickland / Stephen M Soisson / Daniel J Klein / Anthony T Ginnetti / Michael J Marino / Shawn J Stachel / Andrii Ishchenko /  Abstract: Voltage-gated potassium channels (Kv) are tetrameric membrane proteins that provide a highly selective pathway for potassium ions (K) to diffuse across a hydrophobic cell membrane. These unique ...Voltage-gated potassium channels (Kv) are tetrameric membrane proteins that provide a highly selective pathway for potassium ions (K) to diffuse across a hydrophobic cell membrane. These unique voltage-gated cation channels detect changes in membrane potential and, upon activation, help to return the depolarized cell to a resting state during the repolarization stage of each action potential. The Kv3 family of potassium channels is characterized by a high activation potential and rapid kinetics, which play a crucial role for the fast-spiking neuronal phenotype. Mutations in the Kv3.1 channel have been shown to have implications in various neurological diseases like epilepsy and Alzheimer's disease. Moreover, disruptions in neuronal circuitry involving Kv3.1 have been correlated with negative symptoms of schizophrenia. Here, we report the discovery of a novel positive modulator of Kv3.1, investigate its biophysical properties, and determine the cryo-EM structure of the compound in complex with Kv3.1. Structural analysis reveals the molecular determinants of positive modulation in Kv3.1 channels by this class of compounds and provides additional opportunities for rational drug design for the treatment of associated neurological disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28793.map.gz emd_28793.map.gz | 85.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28793-v30.xml emd-28793-v30.xml emd-28793.xml emd-28793.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28793.png emd_28793.png | 111.9 KB | ||
| Filedesc metadata |  emd-28793.cif.gz emd-28793.cif.gz | 6.2 KB | ||
| Others |  emd_28793_half_map_1.map.gz emd_28793_half_map_1.map.gz emd_28793_half_map_2.map.gz emd_28793_half_map_2.map.gz | 84 MB 84 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28793 http://ftp.pdbj.org/pub/emdb/structures/EMD-28793 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28793 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28793 | HTTPS FTP |
-Related structure data
| Related structure data |  8f1cMC  8f1dC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28793.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28793.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28793_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28793_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : potassium voltage-gated channel
| Entire | Name: potassium voltage-gated channel Voltage-gated potassium channel Voltage-gated potassium channel |
|---|---|
| Components |
|
-Supramolecule #1: potassium voltage-gated channel
| Supramolecule | Name: potassium voltage-gated channel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 240 KDa |
-Macromolecule #1: Potassium voltage-gated channel subfamily C member 1
| Macromolecule | Name: Potassium voltage-gated channel subfamily C member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 62.745266 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADYKDHDGD YKDHDIDYKD DDDKLEVLFQ GPGSMGQGDE SERIVINVGG TRHQTYRSTL RTLPGTRLAW LAEPDAHSHF DYDPRADEF FFDRHPGVFA HILNYYRTGK LHCPADVCGP LYEEELAFWG IDETDVEPCC WMTYRQHRDA EEALDSFGGA P LDNSADDA ...String: MADYKDHDGD YKDHDIDYKD DDDKLEVLFQ GPGSMGQGDE SERIVINVGG TRHQTYRSTL RTLPGTRLAW LAEPDAHSHF DYDPRADEF FFDRHPGVFA HILNYYRTGK LHCPADVCGP LYEEELAFWG IDETDVEPCC WMTYRQHRDA EEALDSFGGA P LDNSADDA DADGPGDSGD GEDELEMTKR LALSDSPDGR PGGFWRRWQP RIWALFEDPY SSRYARYVAF ASLFFILVSI TT FCLETHE RFNPIVNKTE IENVRNGTQV RYYREAETEA FLTYIEGVCV VWFTFEFLMR VIFCPNKVEF IKNSLNIIDF VAI LPFYLE VGLSGLSSKA AKDVLGFLRV VRFVRILRIF KLTRHFVGLR VLGHTLRAST NEFLLLIIFL ALGVLIFATM IYYA ERIGA QPNDPSASEH THFKNIPIGF WWAVVTMTTL GYGDMYPQTW SGMLVGALCA LAGVLTIAMP VPVIVNNFGM YYSLA MAKQ KLPKKKKKHI PRPPQLGSPN YCKSVVNSPH HSTQSDTCPL AQEEILEINR AGRKPLRGMS ILEVLFQG UniProtKB: Potassium voltage-gated channel subfamily C member 1 |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #3: (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)o...
| Macromolecule | Name: (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one type: ligand / ID: 3 / Number of copies: 4 / Formula: X9T |
|---|---|
| Molecular weight | Theoretical: 437.879 Da |
| Chemical component information | 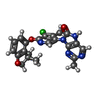 ChemComp-X9T: |
-Macromolecule #4: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 4 / Number of copies: 4 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #5: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 42.5 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 1883143 |
|---|---|
| Startup model | Type of model: OTHER / Details: ab-initio model from cryoSPARC |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.92 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 125905 |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8f1c: |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X

















