+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Microsomal triglyceride transfer protein | |||||||||
 Map data Map data | Microsomal triglyceride transfer protein | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Microsomal triglyceride transfer protein / Microsomal triglyceride transfer protein /  human liver / LIPID TRANSPORT / human liver / LIPID TRANSPORT /  ISOMERASE / ISOMERASE /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information plasma lipoprotein particle assembly / triglyceride transfer activity / plasma lipoprotein particle assembly / triglyceride transfer activity /  chylomicron assembly / phosphatidylcholine transfer activity / regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / procollagen-proline 4-dioxygenase complex / chylomicron assembly / phosphatidylcholine transfer activity / regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / procollagen-proline 4-dioxygenase complex /  VLDL assembly / insulin processing / triglyceride transport / phospholipid transfer activity ... VLDL assembly / insulin processing / triglyceride transport / phospholipid transfer activity ... plasma lipoprotein particle assembly / triglyceride transfer activity / plasma lipoprotein particle assembly / triglyceride transfer activity /  chylomicron assembly / phosphatidylcholine transfer activity / regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / procollagen-proline 4-dioxygenase complex / chylomicron assembly / phosphatidylcholine transfer activity / regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / procollagen-proline 4-dioxygenase complex /  VLDL assembly / insulin processing / triglyceride transport / phospholipid transfer activity / phosphatidylethanolamine transfer activity / procollagen-proline 4-dioxygenase activity / interleukin-23-mediated signaling pathway / LDL remodeling / VLDL assembly / insulin processing / triglyceride transport / phospholipid transfer activity / phosphatidylethanolamine transfer activity / procollagen-proline 4-dioxygenase activity / interleukin-23-mediated signaling pathway / LDL remodeling /  thiol oxidase activity / thiol oxidase activity /  protein disulfide-isomerase / ceramide 1-phosphate transfer activity / phospholipid transporter activity / peptidyl-proline hydroxylation to 4-hydroxy-L-proline / very-low-density lipoprotein particle assembly / endoplasmic reticulum chaperone complex / protein disulfide-isomerase / ceramide 1-phosphate transfer activity / phospholipid transporter activity / peptidyl-proline hydroxylation to 4-hydroxy-L-proline / very-low-density lipoprotein particle assembly / endoplasmic reticulum chaperone complex /  protein folding in endoplasmic reticulum / Collagen biosynthesis and modifying enzymes / protein folding in endoplasmic reticulum / Collagen biosynthesis and modifying enzymes /  Chylomicron assembly / lipid transporter activity / lipoprotein metabolic process / cholesterol transfer activity / phospholipid transport / Interleukin-23 signaling / low-density lipoprotein particle remodeling / interleukin-12-mediated signaling pathway / cellular response to interleukin-7 / Interleukin-12 signaling / lipoprotein transport / triglyceride metabolic process / microvillus membrane / Chylomicron assembly / lipid transporter activity / lipoprotein metabolic process / cholesterol transfer activity / phospholipid transport / Interleukin-23 signaling / low-density lipoprotein particle remodeling / interleukin-12-mediated signaling pathway / cellular response to interleukin-7 / Interleukin-12 signaling / lipoprotein transport / triglyceride metabolic process / microvillus membrane /  protein disulfide isomerase activity / Insulin processing / Detoxification of Reactive Oxygen Species / protein disulfide isomerase activity / Insulin processing / Detoxification of Reactive Oxygen Species /  apolipoprotein binding / apolipoprotein binding /  protein secretion / protein secretion /  protein-disulfide reductase activity / positive regulation of cell adhesion / protein-disulfide reductase activity / positive regulation of cell adhesion /  endoplasmic reticulum-Golgi intermediate compartment / endoplasmic reticulum to Golgi vesicle-mediated transport / positive regulation of substrate adhesion-dependent cell spreading / response to endoplasmic reticulum stress / cholesterol homeostasis / establishment of localization in cell / brush border membrane / Hedgehog ligand biogenesis / endoplasmic reticulum-Golgi intermediate compartment / endoplasmic reticulum to Golgi vesicle-mediated transport / positive regulation of substrate adhesion-dependent cell spreading / response to endoplasmic reticulum stress / cholesterol homeostasis / establishment of localization in cell / brush border membrane / Hedgehog ligand biogenesis /  Post-translational protein phosphorylation / lipid metabolic process / response to calcium ion / Post-translational protein phosphorylation / lipid metabolic process / response to calcium ion /  circadian rhythm / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / circadian rhythm / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) /  melanosome / melanosome /  integrin binding / integrin binding /  protein folding / protein folding /  lamellipodium / lamellipodium /  actin binding / cellular response to hypoxia / basolateral plasma membrane / vesicle / positive regulation of viral entry into host cell / actin binding / cellular response to hypoxia / basolateral plasma membrane / vesicle / positive regulation of viral entry into host cell /  receptor complex / receptor complex /  cytoskeleton / protein heterodimerization activity / external side of plasma membrane / cytoskeleton / protein heterodimerization activity / external side of plasma membrane /  endoplasmic reticulum lumen / endoplasmic reticulum lumen /  focal adhesion / focal adhesion /  lipid binding / protein-containing complex binding / lipid binding / protein-containing complex binding /  Golgi apparatus / Golgi apparatus /  enzyme binding / enzyme binding /  endoplasmic reticulum / protein-containing complex / endoplasmic reticulum / protein-containing complex /  RNA binding / extracellular exosome / extracellular region / RNA binding / extracellular exosome / extracellular region /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
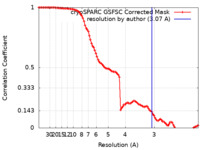
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.07 Å cryo EM / Resolution: 3.07 Å | |||||||||
 Authors Authors | Zhang Z | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: High-resolution structural-omics of human liver enzymes. Authors: Chih-Chia Su / Meinan Lyu / Zhemin Zhang / Masaru Miyagi / Wei Huang / Derek J Taylor / Edward W Yu /  Abstract: We applied raw human liver microsome lysate to a holey carbon grid and used cryo-electron microscopy (cryo-EM) to define its composition. From this sample we identified and simultaneously determined ...We applied raw human liver microsome lysate to a holey carbon grid and used cryo-electron microscopy (cryo-EM) to define its composition. From this sample we identified and simultaneously determined high-resolution structural information for ten unique human liver enzymes involved in diverse cellular processes. Notably, we determined the structure of the endoplasmic bifunctional protein H6PD, where the N- and C-terminal domains independently possess glucose-6-phosphate dehydrogenase and 6-phosphogluconolactonase enzymatic activity, respectively. We also obtained the structure of heterodimeric human GANAB, an ER glycoprotein quality-control machinery that contains a catalytic α subunit and a noncatalytic β subunit. In addition, we observed a decameric peroxidase, PRDX4, which directly contacts a disulfide isomerase-related protein, ERp46. Structural data suggest that several glycosylations, bound endogenous compounds, and ions associate with these human liver enzymes. These results highlight the importance of cryo-EM in facilitating the elucidation of human organ proteomics at the atomic level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28377.map.gz emd_28377.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28377-v30.xml emd-28377-v30.xml emd-28377.xml emd-28377.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28377_fsc.xml emd_28377_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_28377.png emd_28377.png | 96.3 KB | ||
| Filedesc metadata |  emd-28377.cif.gz emd-28377.cif.gz | 5.9 KB | ||
| Others |  emd_28377_additional_1.map.gz emd_28377_additional_1.map.gz emd_28377_half_map_1.map.gz emd_28377_half_map_1.map.gz emd_28377_half_map_2.map.gz emd_28377_half_map_2.map.gz | 52.1 MB 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28377 http://ftp.pdbj.org/pub/emdb/structures/EMD-28377 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28377 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28377 | HTTPS FTP |
-Related structure data
| Related structure data |  8eojMC  7uzmC  8ekwC  8ekyC  8em2C  8emrC  8emsC  8emtC  8eneC  8eorC  23426 M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28377.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28377.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Microsomal triglyceride transfer protein | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Microsomal triglyceride transfer protein
| File | emd_28377_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Microsomal triglyceride transfer protein | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Microsomal triglyceride transfer protein
| File | emd_28377_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Microsomal triglyceride transfer protein | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Microsomal triglyceride transfer protein
| File | emd_28377_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Microsomal triglyceride transfer protein | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Microsomal triglyceride transfer protein
| Entire | Name: Microsomal triglyceride transfer protein |
|---|---|
| Components |
|
-Supramolecule #1: Microsomal triglyceride transfer protein
| Supramolecule | Name: Microsomal triglyceride transfer protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Protein disulfide-isomerase
| Macromolecule | Name: Protein disulfide-isomerase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number:  protein disulfide-isomerase protein disulfide-isomerase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.190137 KDa |
| Sequence | String: MLRRALLCLA VAALVRADAP EEEDHVLVLR KSNFAEALAA HKYLLVEFYA PWCGHCKALA PEYAKAAGKL KAEGSEIRLA KVDATEESD LAQQYGVRGY PTIKFFRNGD TASPKEYTAG READDIVNWL KKRTGPAATT LPDGAAAESL VESSEVAVIG F FKDVESDS ...String: MLRRALLCLA VAALVRADAP EEEDHVLVLR KSNFAEALAA HKYLLVEFYA PWCGHCKALA PEYAKAAGKL KAEGSEIRLA KVDATEESD LAQQYGVRGY PTIKFFRNGD TASPKEYTAG READDIVNWL KKRTGPAATT LPDGAAAESL VESSEVAVIG F FKDVESDS AKQFLQAAEA IDDIPFGITS NSDVFSKYQL DKDGVVLFKK FDEGRNNFEG EVTKENLLDF IKHNQLPLVI EF TEQTAPK IFGGEIKTHI LLFLPKSVSD YDGKLSNFKT AAESFKGKIL FIFIDSDHTD NQRILEFFGL KKEECPAVRL ITL EEEMTK YKPESEELTA ERITEFCHRF LEGKIKPHLM SQELPEDWDK QPVKVLVGKN FEDVAFDEKK NVFVEFYAPW CGHC KQLAP IWDKLGETYK DHENIVIAKM DSTANEVEAV KVHSFPTLKF FPASADRTVI DYNGERTLDG FKKFLESGGQ DGAGD DDDL EDLEEAEEPD MEEDDDQKAV KDEL UniProtKB:  Protein disulfide-isomerase Protein disulfide-isomerase |
-Macromolecule #2: Microsomal triglyceride transfer protein large subunit
| Macromolecule | Name: Microsomal triglyceride transfer protein large subunit type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 99.474102 KDa |
| Sequence | String: MILLAVLFLC FISSYSASVK GHTTGLSLNN DRLYKLTYST EVLLDRGKGK LQDSVGYRIS SNVDVALLWR NPDGDDDQLI QITMKDVNV ENVNQQRGEK SIFKGKSPSK IMGKENLEAL QRPTLLHLIH GKVKEFYSYQ NEAVAIENIK RGLASLFQTQ L SSGTTNEV ...String: MILLAVLFLC FISSYSASVK GHTTGLSLNN DRLYKLTYST EVLLDRGKGK LQDSVGYRIS SNVDVALLWR NPDGDDDQLI QITMKDVNV ENVNQQRGEK SIFKGKSPSK IMGKENLEAL QRPTLLHLIH GKVKEFYSYQ NEAVAIENIK RGLASLFQTQ L SSGTTNEV DISGNCKVTY QAHQDKVIKI KALDSCKIAR SGFTTPNQVL GVSSKATSVT TYKIEDSFVI AVLAEETHNF GL NFLQTIK GKIVSKQKLE LKTTEAGPRL MSGKQAAAII KAVDSKYTAI PIVGQVFQSH CKGCPSLSEL WRSTRKYLQP DNL SKAEAV RNFLAFIQHL RTAKKEEILQ ILKMENKEVL PQLVDAVTSA QTSDSLEAIL DFLDFKSDSS IILQERFLYA CGFA SHPNE ELLRALISKF KGSIGSSDIR ETVMIITGTL VRKLCQNEGC KLKAVVEAKK LILGGLEKAE KKEDTRMYLL ALKNA LLPE GIPSLLKYAE AGEGPISHLA TTALQRYDLP FITDEVKKTL NRIYHQNRKV HEKTVRTAAA AIILNNNPSY MDVKNI LLS IGELPQEMNK YMLAIVQDIL RFEMPASKIV RRVLKEMVAH NYDRFSRSGS SSAYTGYIER SPRSASTYSL DILYSGS GI LRRSNLNIFQ YIGKAGLHGS QVVIEAQGLE ALIAATPDEG EENLDSYAGM SAILFDVQLR PVTFFNGYSD LMSKMLSA S GDPISVVKGL ILLIDHSQEL QLQSGLKANI EVQGGLAIDI SGAMEFSLWY RESKTRVKNR VTVVITTDIT VDSSFVKAG LETSTETEAG LEFISTVQFS QYPFLVCMQM DKDEAPFRQF EKKYERLSTG RGYVSQKRKE SVLAGCEFPL HQENSEMCKV VFAPQPDST SSGWF UniProtKB:  Microsomal triglyceride transfer protein large subunit Microsomal triglyceride transfer protein large subunit |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.291 µm / Nominal defocus min: 0.17 µm / Nominal magnification: 81000 Bright-field microscopy / Nominal defocus max: 3.291 µm / Nominal defocus min: 0.17 µm / Nominal magnification: 81000 |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 41.25 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z
Z Y
Y X
X