+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hexameric IL-21 complex with IL-21R and IL-2Rg | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | IL-21 /  receptor / receptor /  complex / complex /  signaling / signaling /  CYTOKINE CYTOKINE | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
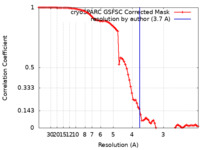
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Abhiraman GC / Jude KM / Caveney NA / Garcia KC | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: A structural blueprint for interleukin-21 signal modulation. Authors: Gita C Abhiraman / Theodora U J Bruun / Nathanael A Caveney / Leon L Su / Robert A Saxton / Qian Yin / Shaogeng Tang / Mark M Davis / Kevin M Jude / K Christopher Garcia /  Abstract: Interleukin-21 (IL-21) plays a critical role in generating immunological memory by promoting the germinal center reaction, yet clinical use of IL-21 remains challenging because of its pleiotropy and ...Interleukin-21 (IL-21) plays a critical role in generating immunological memory by promoting the germinal center reaction, yet clinical use of IL-21 remains challenging because of its pleiotropy and association with autoimmune disease. To better understand the structural basis of IL-21 signaling, we determine the structure of the IL-21-IL-21R-γc ternary signaling complex by X-ray crystallography and a structure of a dimer of trimeric complexes using cryo-electron microscopy. Guided by the structure, we design analogs of IL-21 by introducing substitutions to the IL-21-γc interface. These IL-21 analogs act as partial agonists that modulate downstream activation of pS6, pSTAT3, and pSTAT1. These analogs exhibit differential activity on T and B cell subsets and modulate antibody production in human tonsil organoids. These results clarify the structural basis of IL-21 signaling and offer a potential strategy for tunable manipulation of humoral immunity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28278.map.gz emd_28278.map.gz | 56.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28278-v30.xml emd-28278-v30.xml emd-28278.xml emd-28278.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28278_fsc.xml emd_28278_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_28278.png emd_28278.png | 88.4 KB | ||
| Masks |  emd_28278_msk_1.map emd_28278_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_28278_half_map_1.map.gz emd_28278_half_map_1.map.gz emd_28278_half_map_2.map.gz emd_28278_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28278 http://ftp.pdbj.org/pub/emdb/structures/EMD-28278 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28278 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28278 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28278.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28278.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.151 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28278_msk_1.map emd_28278_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_28278_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_28278_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hexameric IL-21 complex with IL-21R and IL-2Rg
| Entire | Name: Hexameric IL-21 complex with IL-21R and IL-2Rg |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric IL-21 complex with IL-21R and IL-2Rg
| Supramolecule | Name: Hexameric IL-21 complex with IL-21R and IL-2Rg / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Interleukin-21
| Supramolecule | Name: Interleukin-21 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Interleukin-21 receptor
| Supramolecule | Name: Interleukin-21 receptor / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: Cytokine receptor common subunit gamma
| Supramolecule | Name: Cytokine receptor common subunit gamma / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Interleukin-21
| Macromolecule | Name: Interleukin-21 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QGQDRHMIRM RQLIDIVDQL KNYVNDLVPE FLPAPEDVET NCEWSAFSCF QKAQLKSANT GNNERIIQVS IKKLKRKPPS TNAGRRQKHR LTCPSCDSYE KKPPKEFLER FKSLLQKMIH QHLSSRTHGS EDSGAPGSHH HHHH |
-Macromolecule #2: Interleukin-21 receptor
| Macromolecule | Name: Interleukin-21 receptor / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: CPDLVCYTDY LQTVICILEM WNLHPSTLTL TWQDQYEELK DEATSCSLHR SAHNATHATY TCHMDVFHFM ADDIFSVQIT DQSGQYSQEC GSFLLAESIK PAPPFDVTVT FSGQYQISWR SDYEDPAFYM LKGKLQYELQ YRNRGDPWAV SPRRKLISVD SRSVSLLPLE ...String: CPDLVCYTDY LQTVICILEM WNLHPSTLTL TWQDQYEELK DEATSCSLHR SAHNATHATY TCHMDVFHFM ADDIFSVQIT DQSGQYSQEC GSFLLAESIK PAPPFDVTVT FSGQYQISWR SDYEDPAFYM LKGKLQYELQ YRNRGDPWAV SPRRKLISVD SRSVSLLPLE FRKDSSYELQ VRAGPMPGSS YQGTWSEWSD PVIFQTQSEE KAASGRGLND IFEAQKIEWH EHHHHHH |
-Macromolecule #3: Cytokine receptor common subunit gamma
| Macromolecule | Name: Cytokine receptor common subunit gamma / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: PLPLPEVQCF VFNVEYMNCT WQSSSEPQPT NLTLHYWYKN SDNDKVQKCS HYLFSEEITS GCQLQKKEIH LYQTFVVQLQ DPREPRRQAT QMLKLQNLVI PWAPENLTLH KLSESQLELN WNNRFLNHCL EHLVQYRTDW DHSWTEQSVD YRHKFSLPSV DGQKRYTFRV ...String: PLPLPEVQCF VFNVEYMNCT WQSSSEPQPT NLTLHYWYKN SDNDKVQKCS HYLFSEEITS GCQLQKKEIH LYQTFVVQLQ DPREPRRQAT QMLKLQNLVI PWAPENLTLH KLSESQLELN WNNRFLNHCL EHLVQYRTDW DHSWTEQSVD YRHKFSLPSV DGQKRYTFRV RSRFNPLCGS AQHWSEWSHP IHWGSNTSKE NHHHHHH |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 5.0 sec. / Average electron dose: 49.0 e/Å2 |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X