[English] 日本語
 Yorodumi
Yorodumi- EMDB-28075: Structure of Lates calcarifer DNA polymerase theta polymerase dom... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | |||||||||
 Map data Map data | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Lates calcarifer (barramundi perch) Lates calcarifer (barramundi perch) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.4 Å cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Li C / Zhu H / Sun J / Gao Y | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Structural basis of DNA polymerase θ mediated DNA end joining. Authors: Chuxuan Li / Hanwen Zhu / Shikai Jin / Leora M Maksoud / Nikhil Jain / Ji Sun / Yang Gao /  Abstract: DNA polymerase θ (Pol θ) plays an essential role in the microhomology-mediated end joining (MMEJ) pathway for repairing DNA double-strand breaks. However, the mechanisms by which Pol θ recognizes ...DNA polymerase θ (Pol θ) plays an essential role in the microhomology-mediated end joining (MMEJ) pathway for repairing DNA double-strand breaks. However, the mechanisms by which Pol θ recognizes microhomologous DNA ends and performs low-fidelity DNA synthesis remain unclear. Here, we present cryo-electron microscope structures of the polymerase domain of Lates calcarifer Pol θ with long and short duplex DNA at up to 2.4 Å resolution. Interestingly, Pol θ binds to long and short DNA substrates similarly, with extensive interactions around the active site. Moreover, Pol θ shares a similar active site as high-fidelity A-family polymerases with its finger domain well-closed but differs in having hydrophilic residues surrounding the nascent base pair. Computational simulations and mutagenesis studies suggest that the unique insertion loops of Pol θ help to stabilize short DNA binding and assemble the active site for MMEJ repair. Taken together, our results illustrate the structural basis of Pol θ-mediated MMEJ. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28075.map.gz emd_28075.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28075-v30.xml emd-28075-v30.xml emd-28075.xml emd-28075.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28075.png emd_28075.png | 103.1 KB | ||
| Others |  emd_28075_half_map_1.map.gz emd_28075_half_map_1.map.gz emd_28075_half_map_2.map.gz emd_28075_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28075 http://ftp.pdbj.org/pub/emdb/structures/EMD-28075 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28075 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28075 | HTTPS FTP |
-Related structure data
| Related structure data |  8ef9MC  8efcC  8efkC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28075.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28075.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Structure of Lates calcarifer DNA polymerase theta polymerase...
| File | emd_28075_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Structure of Lates calcarifer DNA polymerase theta polymerase...
| File | emd_28075_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
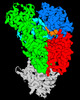
-Entire : Ternary complex of Lates calcarifer DNA polymerase Theta with dup...
| Entire | Name: Ternary complex of Lates calcarifer DNA polymerase Theta with duplex DNA and incoming nucleotide |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of Lates calcarifer DNA polymerase Theta with dup...
| Supramolecule | Name: Ternary complex of Lates calcarifer DNA polymerase Theta with duplex DNA and incoming nucleotide type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Lates calcarifer (barramundi perch) Lates calcarifer (barramundi perch) |
-Macromolecule #1: DNA polymerase theta
| Macromolecule | Name: DNA polymerase theta / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Lates calcarifer (barramundi perch) Lates calcarifer (barramundi perch) |
| Molecular weight | Theoretical: 95.674117 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: MGHHHHHHPS GVKTENNDHI NLKVAGQDGS VVQFKIKRHT PLSKLMKAYC ERQGLSMRQI RFRFDGQPIN ETDTPAQLEM EDEDTIDVF QQQTGGCMDP PSDAESPVTD DGFTLQLSQD ASLCPSNSGT FSIIDVASDR RLFNTFIKEW KTKERYSLAL A CEKREHIQ ...String: MGHHHHHHPS GVKTENNDHI NLKVAGQDGS VVQFKIKRHT PLSKLMKAYC ERQGLSMRQI RFRFDGQPIN ETDTPAQLEM EDEDTIDVF QQQTGGCMDP PSDAESPVTD DGFTLQLSQD ASLCPSNSGT FSIIDVASDR RLFNTFIKEW KTKERYSLAL A CEKREHIQ QPEGEIGGKH KRAPAARQKL NRTDGFPVRD SDGLVLIGLS VCWGARDSYY ISLQQEQSKG LSSSLAPPPL DD DLPVSER LGQVRSCLSR PSAGLRGGVV VTYDIIQVYK TLVLSCGISL AGNCEDPKVA CWLLDPGSEE RTLPNMVTVY CPE ELPLLD GLGSAHAHCP RVRAATKSVL VHAVMNHLTG LLEKDSMLDL FRSIEMPSQV CLALLELNGV GFSVEECERQ KHVM QAKLT ALESQAYNLA GHSFSLTSID DIAQVLFLEL HLPPNGDVGG SKSKKTLGYT RRGGGRVRLG KQFSTTKDIL EKLRP LHPL PGVILEWRRI TNALTKVVFP LQREKQYHPT LAMDRIYPIA QTHTATGRVS FTEPNIQNVP KDFEICMPTV VGESPP SQN GCQMTTKPGK NRRSVAPSVT GGAAEQGPAF SVSMRHAFVP FSGGMILAAD YSQLELRVLA HLSKDQRLLQ VLNGGAD VF RCIAAEWKGV DPETVNDSLR QQAKQICYGI IYGMGAKSLG EQMGVEENDA ACYIESFKAR YKGINAFLKE TVKNCIKN G YVQTLMGRRR YLPGISNTNT HIKAHAERQA VNTTVQGSAA DIVKLATVNI QKRLRKTYPT APLSHQHTHS GTSQYRAGT SHLRGAFFVL QLHDELIYET REEDLIQVAQ IVKREMESAV KLYVKLKAKV KVGPSWGNLQ DLDL |
-Macromolecule #2: DNA (5'-D(*AP*GP*CP*TP*CP*TP*AP*CP*GP*GP*AP*TP*GP*C)-3')
| Macromolecule | Name: DNA (5'-D(*AP*GP*CP*TP*CP*TP*AP*CP*GP*GP*AP*TP*GP*C)-3') type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Lates calcarifer (barramundi perch) Lates calcarifer (barramundi perch) |
| Molecular weight | Theoretical: 8.864712 KDa |
| Sequence | String: (DG)(DC)(DA)(DG)(DT)(DC)(DA)(DG)(DC)(DT) (DC)(DT)(DA)(DC)(DG)(DG)(DA)(DT)(DG)(DC) (DC)(DT)(DC)(DA)(DC)(DA)(DG)(DC)(DA) |
-Macromolecule #3: DNA (5'-D(*AP*GP*CP*AP*TP*CP*CP*GP*TP*AP*GP*(2DA))-3')
| Macromolecule | Name: DNA (5'-D(*AP*GP*CP*AP*TP*CP*CP*GP*TP*AP*GP*(2DA))-3') type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Lates calcarifer (barramundi perch) Lates calcarifer (barramundi perch) |
| Molecular weight | Theoretical: 5.844796 KDa |
| Sequence | String: (DT)(DG)(DC)(DT)(DG)(DT)(DG)(DA)(DG)(DC) (DA)(DT)(DC)(DC)(DG)(DT)(DA)(DG)(2DA) |
-Macromolecule #4: 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: DGT |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-DGT: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.2 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 825204 |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X