+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Local refinement of LRRK1 around the ROC-COR-kinase domains | |||||||||||||||
 Map data Map data | Local refinement around ROC-COR-kinase domains of LRRK1 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords |  monomer / monomer /  TRANSFERASE TRANSFERASE | |||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||||||||
 Authors Authors | Reimer JM / Mathea S / Knapp S / Leschziner AE | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structure of LRRK1 and mechanisms of autoinhibition and activation. Authors: Janice M Reimer / Andrea M Dickey / Yu Xuan Lin / Robert G Abrisch / Sebastian Mathea / Deep Chatterjee / Elizabeth J Fay / Stefan Knapp / Matthew D Daugherty / Samara L Reck-Peterson / Andres E Leschziner /   Abstract: Leucine Rich Repeat Kinase 1 and 2 (LRRK1 and LRRK2) are homologs in the ROCO family of proteins in humans. Despite their shared domain architecture and involvement in intracellular trafficking, ...Leucine Rich Repeat Kinase 1 and 2 (LRRK1 and LRRK2) are homologs in the ROCO family of proteins in humans. Despite their shared domain architecture and involvement in intracellular trafficking, their disease associations are strikingly different: LRRK2 is involved in familial Parkinson's disease while LRRK1 is linked to bone diseases. Furthermore, Parkinson's disease-linked mutations in LRRK2 are typically autosomal dominant gain-of-function while those in LRRK1 are autosomal recessive loss-of-function. Here, to understand these differences, we solved cryo-EM structures of LRRK1 in its monomeric and dimeric forms. Both differ from the corresponding LRRK2 structures. Unlike LRRK2, which is sterically autoinhibited as a monomer, LRRK1 is sterically autoinhibited in a dimer-dependent manner. LRRK1 has an additional level of autoinhibition that prevents activation of the kinase and is absent in LRRK2. Finally, we place the structural signatures of LRRK1 and LRRK2 in the context of the evolution of the LRRK family of proteins. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27815.map.gz emd_27815.map.gz | 86.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27815-v30.xml emd-27815-v30.xml emd-27815.xml emd-27815.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27815_fsc.xml emd_27815_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_27815.png emd_27815.png | 31.9 KB | ||
| Filedesc metadata |  emd-27815.cif.gz emd-27815.cif.gz | 6 KB | ||
| Others |  emd_27815_half_map_1.map.gz emd_27815_half_map_1.map.gz emd_27815_half_map_2.map.gz emd_27815_half_map_2.map.gz | 84.6 MB 84.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27815 http://ftp.pdbj.org/pub/emdb/structures/EMD-27815 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27815 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27815 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27815.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27815.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement around ROC-COR-kinase domains of LRRK1 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Local refinement around ROC-COR-kinase domains of LRRK1, Half map A
| File | emd_27815_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement around ROC-COR-kinase domains of LRRK1, Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
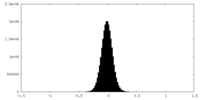
| Density Histograms |
-Half map: Local refinement around ROC-COR-kinase domains of LRRK1, Half map B
| File | emd_27815_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement around ROC-COR-kinase domains of LRRK1, Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
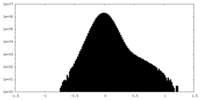
| Density Histograms |
- Sample components
Sample components
-Entire : Monomeric structure of LRRK1
| Entire | Name: Monomeric structure of LRRK1 |
|---|---|
| Components |
|
-Supramolecule #1: Monomeric structure of LRRK1
| Supramolecule | Name: Monomeric structure of LRRK1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Leucine-rich repeat kinase 1
| Macromolecule | Name: Leucine-rich repeat kinase 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: SAVCPERAME TLNGAGDTGG KPSTRGGDPA ARSRRTEGIR AAYRRGDRGG ARDLLEEACD QCASQLEKGQ LLSIPAAYGD LEMVRYLLSK RLVELPTEPT DDNPAVVAAY FGHTAVVQEL LESLPGPCSP QRLLNWMLAL ACQRGHLGVV KLLVLTHGAD PESYAVRKNE ...String: SAVCPERAME TLNGAGDTGG KPSTRGGDPA ARSRRTEGIR AAYRRGDRGG ARDLLEEACD QCASQLEKGQ LLSIPAAYGD LEMVRYLLSK RLVELPTEPT DDNPAVVAAY FGHTAVVQEL LESLPGPCSP QRLLNWMLAL ACQRGHLGVV KLLVLTHGAD PESYAVRKNE FPVIVRLPLY AAIKSGNEDI AIFLLRHGAY FCSYILLDSP DPSKHLLRKY FIEASPLPSS YPGKTALRVK WSHLRLPWVD LDWLIDISCQ ITELDLSANC LATLPSVIPW GLINLRKLNL SDNHLGELPG VQSSDEIICS RLLEIDISSN KLSHLPPGFL HLSKLQKLTA SKNCLEKLFE EENATNWIGL RKLQELDISD NKLTELPALF LHSFKSLNSL NVSRNNLKVF PDPWACPLKC CKASRNALEC LPDKMAVFWK NHLKDVDFSE NALKEVPLGL FQLDALMFLR LQGNQLAALP PQEKWTCRQL KTLDLSRNQL GKNEDGLKTK RIAFFTTRGR QRSGTEAASV LEFPAFLSES LEVLCLNDNH LDTVPPSVCL LKSLSELYLG NNPGLRELPP ELGQLGNLWQ LDTEDLTISN VPAEIQKEGP KAMLSYLRAQ LRKAEKCKLM KMIIVGPPRQ GKSTLLEILQ TGRAPQVVHG EATIRTTKWE LQRPAGSRAK VESVEFNVWD IGGPASMATV NQCFFTDKAL YVVVWNLALG EEAVANLQFW LLNIEAKAPN AVVLVVGTHL DLIEAKFRVE RIATLRAYVL ALCRSPSGSR ATGFPDITFK HLHEISCKSL EGQEGLRQLI FHVTCSMKDV GSTIGCQRLA GRLIPRSYLS LQEAVLAEQQ RRSRDDDVQY LTDRQLEQLV EQTPDNDIKD YEDLQSAISF LIETGTLLHF PDTSHGLRNL YFLDPIWLSE CLQRIFNIKG SRSVAKNGVI RAEDLRMLLV GTGFTQQTEE QYFQFLAKFE IALPVANDSY LLPHLLPSKP GLDTHGMRHP TANTIQRVFK MSFVPVGFWQ RFIARMLISL AEMDLQLFEN KKNTKSRNRK VTIYSFTGNQ RNRCSTFRVK RNQTIYWQEG LLVTFDGGYL SVESSDVNWK KKKSGGMKIV CQSEVRDFSA MAFITDHVNS LIDQWFPALT ATESDGTPLM EQYVPCPVCE TAWAQHTDPS EKSEDVQYFD MEDCVLTAIE RDFISCPRHP DLPVPLQELV PELFMTDFPA RLFLENSKLE HSEDEGSVLG QGGSGTVIYR ARYQGQPVAV KRFHIKKFKN FANVPADTML RHLRATDAMK NFSEFRQEAS MLHALQHPCI VALIGISIHP LCFALELAPL SSLNTVLSEN ARDSSFIPLG HMLTQKIAYQ IASGLAYLHK KNIIFCDLKS DNILVWSLDV KEHINIKLSD YGISRQSFHE GALGVEGTPG YQAPEIRPRI VYDEKVDMFS YGMVLYELLS GQRPALGHHQ LQIAKKLSKG IRPVLGQPEE VQFRRLQALM MECWDTKPEK RPLALSVVSQ MKDPTFATFM YELCCGKQTA FFSSQGQEYT VVFWDGKEES RNYTVVNTEK GLMEVQRMCC PGMKVSCQLQ VQRSLWTATE DQKIYIYTLK GMCPLNTPQQ ALDTPAVVTC FLAVPVIKKN SYLVLAGLAD GLVAVFPVVR GTPKDSCSYL CSHTANRSKF SIADEDARQN PYPVKAMEVV NSGSEVWYSN GPGLLVIDCA SLEICRRLEP YMAPSMVTSV VCSSEGRGEE VVWCLDDKAN SLVMYHSTTY QLCARYFCGV PSPLRDMFPV RPLDTEPPAA SHTANPKVPE GDSIADVSIM YSEELGTQIL IHQESLTDYC SMSSYSSSPP RQAARSPSSL PSSPASSSSV PFSTDCEDSD MLHTPGAASD RSEHDLTPMD GETFSQHLQA VKILAVRDLI WVPRRGGDVI VIGLEKDSGA QRGRVIAVLK ARELTPHGVL VDAAVVAKDT VVCTFENENT EWCLAVWRGW GAREFDIFYQ SYEELGRLEA CTRKRR |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: 50 mM HEPES pH 7.4, 150 mM NaCl, 5% glycerol, 0.5 mM TCEP, 20 uM GDP |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.124 µm / Nominal defocus min: 1.2630000000000001 µm Bright-field microscopy / Nominal defocus max: 2.124 µm / Nominal defocus min: 1.2630000000000001 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 51.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X