[English] 日本語
 Yorodumi
Yorodumi- EMDB-27642: The structure of the IL-11 signalling complex, with full-length e... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
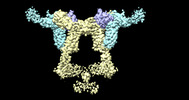
| Title | The structure of the IL-11 signalling complex, with full-length extracellular gp130 | |||||||||
 Map data Map data | The structure of the IL-11 signalling complex, with full-length extracellular gp130 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Complex / GP130 / glycoprotein 130 / Complex / GP130 / glycoprotein 130 /  signalling / signalling /  cancer / cancer /  CYTOKINE CYTOKINE | |||||||||
| Function / homology |  Function and homology information Function and homology information interleukin-11 receptor binding / interleukin-11 receptor binding /  interleukin-27 receptor activity / oncostatin-M receptor complex / oncostatin-M-mediated signaling pathway / interleukin-27 receptor activity / oncostatin-M receptor complex / oncostatin-M-mediated signaling pathway /  ciliary neurotrophic factor receptor activity / ciliary neurotrophic factor receptor activity /  ciliary neurotrophic factor receptor binding / negative regulation of interleukin-6-mediated signaling pathway / leukemia inhibitory factor signaling pathway / ciliary neurotrophic factor receptor binding / negative regulation of interleukin-6-mediated signaling pathway / leukemia inhibitory factor signaling pathway /  interleukin-11 receptor activity / interleukin-11 receptor activity /  interleukin-11 binding ... interleukin-11 binding ... interleukin-11 receptor binding / interleukin-11 receptor binding /  interleukin-27 receptor activity / oncostatin-M receptor complex / oncostatin-M-mediated signaling pathway / interleukin-27 receptor activity / oncostatin-M receptor complex / oncostatin-M-mediated signaling pathway /  ciliary neurotrophic factor receptor activity / ciliary neurotrophic factor receptor activity /  ciliary neurotrophic factor receptor binding / negative regulation of interleukin-6-mediated signaling pathway / leukemia inhibitory factor signaling pathway / ciliary neurotrophic factor receptor binding / negative regulation of interleukin-6-mediated signaling pathway / leukemia inhibitory factor signaling pathway /  interleukin-11 receptor activity / interleukin-11 receptor activity /  interleukin-11 binding / megakaryocyte differentiation / interleukin-11 binding / megakaryocyte differentiation /  ciliary neurotrophic factor receptor complex / interleukin-27-mediated signaling pathway / ciliary neurotrophic factor-mediated signaling pathway / ciliary neurotrophic factor receptor complex / interleukin-27-mediated signaling pathway / ciliary neurotrophic factor-mediated signaling pathway /  interleukin-6 receptor complex / head development / negative regulation of hormone secretion / interleukin-11-mediated signaling pathway / T-helper 17 cell lineage commitment / positive regulation of adaptive immune response / interleukin-6 receptor complex / head development / negative regulation of hormone secretion / interleukin-11-mediated signaling pathway / T-helper 17 cell lineage commitment / positive regulation of adaptive immune response /  developmental process / positive regulation of acute inflammatory response / positive regulation of astrocyte differentiation / intestinal epithelial cell development / positive regulation of platelet aggregation / Interleukin-35 Signalling / Interleukin-27 signaling / IL-6-type cytokine receptor ligand interactions / developmental process / positive regulation of acute inflammatory response / positive regulation of astrocyte differentiation / intestinal epithelial cell development / positive regulation of platelet aggregation / Interleukin-35 Signalling / Interleukin-27 signaling / IL-6-type cytokine receptor ligand interactions /  cytokine receptor activity / glycogen metabolic process / Interleukin-6 signaling / interleukin-6-mediated signaling pathway / positive regulation of Notch signaling pathway / protein tyrosine kinase activator activity / cytokine receptor activity / glycogen metabolic process / Interleukin-6 signaling / interleukin-6-mediated signaling pathway / positive regulation of Notch signaling pathway / protein tyrosine kinase activator activity /  cytokine binding / positive regulation of cardiac muscle hypertrophy / fat cell differentiation / MAPK3 (ERK1) activation / cytokine binding / positive regulation of cardiac muscle hypertrophy / fat cell differentiation / MAPK3 (ERK1) activation /  growth factor binding / MAPK1 (ERK2) activation / positive regulation of vascular endothelial growth factor production / positive regulation of osteoblast differentiation / growth factor binding / MAPK1 (ERK2) activation / positive regulation of vascular endothelial growth factor production / positive regulation of osteoblast differentiation /  coreceptor activity / positive regulation of tyrosine phosphorylation of STAT protein / positive regulation of T cell proliferation / B cell differentiation / response to cytokine / coreceptor activity / positive regulation of tyrosine phosphorylation of STAT protein / positive regulation of T cell proliferation / B cell differentiation / response to cytokine /  cytokine activity / cytokine activity /  growth factor activity / cytokine-mediated signaling pathway / positive regulation of peptidyl-tyrosine phosphorylation / transmembrane signaling receptor activity / positive regulation of peptidyl-serine phosphorylation / growth factor activity / cytokine-mediated signaling pathway / positive regulation of peptidyl-tyrosine phosphorylation / transmembrane signaling receptor activity / positive regulation of peptidyl-serine phosphorylation /  scaffold protein binding / cell population proliferation / negative regulation of neuron apoptotic process / positive regulation of MAPK cascade / scaffold protein binding / cell population proliferation / negative regulation of neuron apoptotic process / positive regulation of MAPK cascade /  receptor complex / receptor complex /  membrane raft / external side of plasma membrane / neuronal cell body / membrane raft / external side of plasma membrane / neuronal cell body /  dendrite / positive regulation of cell population proliferation / negative regulation of apoptotic process / positive regulation of transcription by RNA polymerase II / dendrite / positive regulation of cell population proliferation / negative regulation of apoptotic process / positive regulation of transcription by RNA polymerase II /  extracellular space / extracellular exosome / extracellular region / extracellular space / extracellular exosome / extracellular region /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
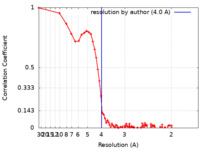
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.0 Å cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Metcalfe RD / Hanssen E / Griffin MDW | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant. Authors: Riley D Metcalfe / Eric Hanssen / Ka Yee Fung / Kaheina Aizel / Clara C Kosasih / Courtney O Zlatic / Larissa Doughty / Craig J Morton / Andrew P Leis / Michael W Parker / Paul R Gooley / ...Authors: Riley D Metcalfe / Eric Hanssen / Ka Yee Fung / Kaheina Aizel / Clara C Kosasih / Courtney O Zlatic / Larissa Doughty / Craig J Morton / Andrew P Leis / Michael W Parker / Paul R Gooley / Tracy L Putoczki / Michael D W Griffin /   Abstract: Interleukin (IL-)11, an IL-6 family cytokine, has pivotal roles in autoimmune diseases, fibrotic complications, and solid cancers. Despite intense therapeutic targeting efforts, structural ...Interleukin (IL-)11, an IL-6 family cytokine, has pivotal roles in autoimmune diseases, fibrotic complications, and solid cancers. Despite intense therapeutic targeting efforts, structural understanding of IL-11 signalling and mechanistic insights into current inhibitors are lacking. Here we present cryo-EM and crystal structures of the human IL-11 signalling complex, including the complex containing the complete extracellular domains of the shared IL-6 family β-receptor, gp130. We show that complex formation requires conformational reorganisation of IL-11 and that the membrane-proximal domains of gp130 are dynamic. We demonstrate that the cytokine mutant, IL-11 Mutein, competitively inhibits signalling in human cell lines. Structural shifts in IL-11 Mutein underlie inhibition by altering cytokine binding interactions at all three receptor-engaging sites and abrogating the final gp130 binding step. Our results reveal the structural basis of IL-11 signalling, define the molecular mechanisms of an inhibitor, and advance understanding of gp130-containing receptor complexes, with potential applications in therapeutic development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27642.map.gz emd_27642.map.gz | 93 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27642-v30.xml emd-27642-v30.xml emd-27642.xml emd-27642.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27642_fsc.xml emd_27642_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_27642.png emd_27642.png | 53.1 KB | ||
| Filedesc metadata |  emd-27642.cif.gz emd-27642.cif.gz | 6.6 KB | ||
| Others |  emd_27642_half_map_1.map.gz emd_27642_half_map_1.map.gz emd_27642_half_map_2.map.gz emd_27642_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27642 http://ftp.pdbj.org/pub/emdb/structures/EMD-27642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27642 | HTTPS FTP |
-Related structure data
| Related structure data |  8dptMC  8dpsC  8dpuC  8dpvC  8dpwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27642.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27642.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The structure of the IL-11 signalling complex, with full-length extracellular gp130 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: The structure of the IL-11 signalling complex, with...
| File | emd_27642_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The structure of the IL-11 signalling complex, with full-length extracellular gp130 | ||||||||||||
| Projections & Slices |
| ||||||||||||
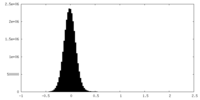
| Density Histograms |
-Half map: The structure of the IL-11 signalling complex, with...
| File | emd_27642_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The structure of the IL-11 signalling complex, with full-length extracellular gp130 | ||||||||||||
| Projections & Slices |
| ||||||||||||
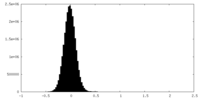
| Density Histograms |
- Sample components
Sample components
-Entire : Interleukin 11 signalling complex
| Entire | Name: Interleukin 11 signalling complex |
|---|---|
| Components |
|
-Supramolecule #1: Interleukin 11 signalling complex
| Supramolecule | Name: Interleukin 11 signalling complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: 2:2:2 hexameric complex of IL-11, IL-11Ra and gp130. Purified from recombinantly expressed and purified proteins. |
|---|---|
| Molecular weight | Theoretical: 290 KDa |
-Supramolecule #2: gp130
| Supramolecule | Name: gp130 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: IL-11Ra
| Supramolecule | Name: IL-11Ra / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: IL-11
| Supramolecule | Name: IL-11 / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Interleukin-6 receptor subunit beta
| Macromolecule | Name: Interleukin-6 receptor subunit beta / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.199836 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: GELLDPCGYI SPESPVVQLH SNFTAVCVLK EKCMDYFHVN ANYIVWKTNH FTIPKEQYTI INRTASSVTF TDIASLNIQL TCNILTFGQ LEQNVYGITI ISGLPPEKPK NLSCIVNEGK KMRCEWDGGR ETHLETNFTL KSEWATHKFA DCKAKRDTPT S CTVDYSTV ...String: GELLDPCGYI SPESPVVQLH SNFTAVCVLK EKCMDYFHVN ANYIVWKTNH FTIPKEQYTI INRTASSVTF TDIASLNIQL TCNILTFGQ LEQNVYGITI ISGLPPEKPK NLSCIVNEGK KMRCEWDGGR ETHLETNFTL KSEWATHKFA DCKAKRDTPT S CTVDYSTV YFVNIEVWVE AENALGKVTS DHINFDPVYK VKPNPPHNLS VINSEELSSI LKLTWTNPSI KSVIILKYNI QY RTKDAST WSQIPPEDTA STRSSFTVQD LKPFTEYVFR IRCMKEDGKG YWSDWSEEAS GITYEDRPSK APSFWYKIDP SHT QGYRTV QLVWKTLPPF EANGKILDYE VTLTRWKSHL QNYTVNATKL TVNLTNDRYL ATLTVRNLVG KSDAAVLTIP ACDF QATHP VMDLKAFPKD NMLWVEWTTP RESVKKYILE WCVLSDKAPC ITDWQQEDGT VHRTYLRGNL AESKCYLITV TPVYA DGPG SPESIKAYLK QAPPSKGPTV RTKKVGKNEA VLEWDQLPVD VQNGFIRNYT IFYRTIIGNE TAVNVDSSHT EYTLSS LTS DTLYMVRMAA YTDEGGKDGP EFTFTTPK UniProtKB:  Interleukin-6 receptor subunit beta Interleukin-6 receptor subunit beta |
-Macromolecule #2: Interleukin-11
| Macromolecule | Name: Interleukin-11 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.273289 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21 (bacteria) Escherichia coli BL21 (bacteria) |
| Sequence | String: GSPDPRAELD STVLLTRSLL ADTRQLAAQL RDKFPADGDH NLDSLPTLAM SAGALGALQL PGVLTRLRAD LLSYLRHVQW LRRAGGSSL KTLEPELGTL QARLDRLLRR LQLLMSRLAL PQPPPDPPAP PLAPPSSAWG GIRAAHAILG GLHLTLDWAV R GLLLLKTR L UniProtKB:  Interleukin-11 Interleukin-11 |
-Macromolecule #3: Interleukin-11 receptor subunit alpha
| Macromolecule | Name: Interleukin-11 receptor subunit alpha / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.208002 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: GSSPCPQAWG PPGVQYGQPG RSVKLCCPGV TAGDPVSWFR DGEPKLLQGP DSGLGHELVL AQADSTDEGT YICQTLDGAL GGTVTLQLG YPPARPVVSC QAADYENFSC TWSPSQISGL PTRYLTSYRK KTVLGADSQR RSPSTGPWPC PQDPLGAARC V VHGAEFWS ...String: GSSPCPQAWG PPGVQYGQPG RSVKLCCPGV TAGDPVSWFR DGEPKLLQGP DSGLGHELVL AQADSTDEGT YICQTLDGAL GGTVTLQLG YPPARPVVSC QAADYENFSC TWSPSQISGL PTRYLTSYRK KTVLGADSQR RSPSTGPWPC PQDPLGAARC V VHGAEFWS QYRINVTEVN PLGASTRLLD VSLQSILRPD PPQGLRVESV PGYPRRLRAS WTYPASWPSQ PHFLLKFRLQ YR PAQHPAW STVEPAGLEE VITDAVAGLP HAVRVSARDF LDAGTWSTWS PEAWGTPSTG T UniProtKB:  Interleukin-11 receptor subunit alpha Interleukin-11 receptor subunit alpha |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X