[English] 日本語
 Yorodumi
Yorodumi- EMDB-26936: Complex of Plasmodium falciparum circumsporozoite protein with 850 Fab -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex of Plasmodium falciparum circumsporozoite protein with 850 Fab | |||||||||||||||||||||
 Map data Map data | map of Plasmodium falciparum CSP in complex with 850 Fab. | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell surface binding / symbiont entry into host / entry into host cell by a symbiont-containing vacuole /  heparan sulfate proteoglycan binding / side of membrane / heparan sulfate proteoglycan binding / side of membrane /  cell surface / cell surface /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||||||||||||||
| Biological species |   Plasmodium falciparum (malaria parasite P. falciparum) / Plasmodium falciparum (malaria parasite P. falciparum) /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||||||||||||||
 Authors Authors | Kucharska I / Prieto K / Rubinstein JL / Julien JP | |||||||||||||||||||||
| Funding support |  Canada, Canada,  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2022 Journal: PLoS Pathog / Year: 2022Title: High-density binding to Plasmodium falciparum circumsporozoite protein repeats by inhibitory antibody elicited in mouse with human immunoglobulin repertoire. Authors: Iga Kucharska / Špela Binter / Rajagopal Murugan / Stephen W Scally / Julia Ludwig / Katherine Prieto / Elaine Thai / Giulia Costa / Kan Li / Gillian Q Horn / Yevel Flores-Garcia / ...Authors: Iga Kucharska / Špela Binter / Rajagopal Murugan / Stephen W Scally / Julia Ludwig / Katherine Prieto / Elaine Thai / Giulia Costa / Kan Li / Gillian Q Horn / Yevel Flores-Garcia / Alexandre Bosch / Taylor Sicard / John L Rubinstein / Fidel Zavala / S Moses Dennison / Georgia D Tomaras / Elena A Levashina / Paul Kellam / Hedda Wardemann / Jean-Philippe Julien /     Abstract: Antibodies targeting the human malaria parasite Plasmodium falciparum circumsporozoite protein (PfCSP) can prevent infection and disease. PfCSP contains multiple central repeating NANP motifs; some ...Antibodies targeting the human malaria parasite Plasmodium falciparum circumsporozoite protein (PfCSP) can prevent infection and disease. PfCSP contains multiple central repeating NANP motifs; some of the most potent anti-infective antibodies against malaria bind to these repeats. Multiple antibodies can bind the repeating epitopes concurrently by engaging into homotypic Fab-Fab interactions, which results in the ordering of the otherwise largely disordered central repeat into a spiral. Here, we characterize IGHV3-33/IGKV1-5-encoded monoclonal antibody (mAb) 850 elicited by immunization of transgenic mice with human immunoglobulin loci. mAb 850 binds repeating NANP motifs with picomolar affinity, potently inhibits Plasmodium falciparum (Pf) in vitro and, when passively administered in a mouse challenge model, reduces liver burden to a similar extent as some of the most potent anti-PfCSP mAbs yet described. Like other IGHV3-33/IGKV1-5-encoded anti-NANP antibodies, mAb 850 primarily utilizes its HCDR3 and germline-encoded aromatic residues to recognize its core NANP motif. Biophysical and cryo-electron microscopy analyses reveal that up to 19 copies of Fab 850 can bind the PfCSP repeat simultaneously, and extensive homotypic interactions are observed between densely-packed PfCSP-bound Fabs to indirectly improve affinity to the antigen. Together, our study expands on the molecular understanding of repeat-induced homotypic interactions in the B cell response against PfCSP for potently protective mAbs against Pf infection. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26936.map.gz emd_26936.map.gz | 141.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26936-v30.xml emd-26936-v30.xml emd-26936.xml emd-26936.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26936.png emd_26936.png | 104.9 KB | ||
| Others |  emd_26936_half_map_1.map.gz emd_26936_half_map_1.map.gz emd_26936_half_map_2.map.gz emd_26936_half_map_2.map.gz | 139 MB 139 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26936 http://ftp.pdbj.org/pub/emdb/structures/EMD-26936 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26936 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26936 | HTTPS FTP |
-Related structure data
| Related structure data |  7v05MC  7uylC  7uymC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26936.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26936.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map of Plasmodium falciparum CSP in complex with 850 Fab. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map of Plasmodium falciparum CSP in complex with 850 Fab.
| File | emd_26936_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of Plasmodium falciparum CSP in complex with 850 Fab. | ||||||||||||
| Projections & Slices |
| ||||||||||||
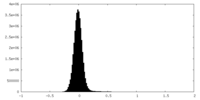
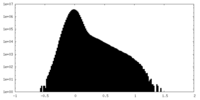
| Density Histograms |
-Half map: half map of Plasmodium falciparum CSP in complex with 850 Fab.
| File | emd_26936_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of Plasmodium falciparum CSP in complex with 850 Fab. | ||||||||||||
| Projections & Slices |
| ||||||||||||
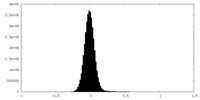
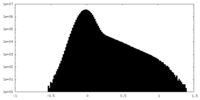
| Density Histograms |
- Sample components
Sample components
-Entire : complex of Plasmodium falciparum circumsporozoite protein with 85...
| Entire | Name: complex of Plasmodium falciparum circumsporozoite protein with 850 Fab. |
|---|---|
| Components |
|
-Supramolecule #1: complex of Plasmodium falciparum circumsporozoite protein with 85...
| Supramolecule | Name: complex of Plasmodium falciparum circumsporozoite protein with 850 Fab. type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Plasmodium falciparum circumsporozoite protein
| Supramolecule | Name: Plasmodium falciparum circumsporozoite protein / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:   Plasmodium falciparum (malaria parasite P. falciparum) Plasmodium falciparum (malaria parasite P. falciparum) |
-Supramolecule #3: 850 Fab
| Supramolecule | Name: 850 Fab / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Macromolecule #1: 850 Fab Heavy Chain
| Macromolecule | Name: 850 Fab Heavy Chain / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 24.234037 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVESGGG VVQPGRSLRL SCAASGFTFS NFGMHWIRQS PGKGLEWVAI IWYDGSNTYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCAKV WFGESEDNYS VDVWGQGTTV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: QVQLVESGGG VVQPGRSLRL SCAASGFTFS NFGMHWIRQS PGKGLEWVAI IWYDGSNTYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCAKV WFGESEDNYS VDVWGQGTTV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSC |
-Macromolecule #2: 850 Fab Light Chain
| Macromolecule | Name: 850 Fab Light Chain / type: protein_or_peptide / ID: 2 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 23.476045 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPST LSTSVGDRVT ITCRASQSIS NWLAWYQQKP GKAPKLLIYK ASTLESGVPS RFSGSGSGTE FTLTISSLQP DDFATYYCQ QYSSYWTFGQ GTKLEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ E SVTEQDSK ...String: DIQMTQSPST LSTSVGDRVT ITCRASQSIS NWLAWYQQKP GKAPKLLIYK ASTLESGVPS RFSGSGSGTE FTLTISSLQP DDFATYYCQ QYSSYWTFGQ GTKLEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ E SVTEQDSK DSTYSLSSTL TLSKADYEKH KVYACEVTHQ GLSSPVTKSF NRGEC |
-Macromolecule #3: Circumsporozoite protein
| Macromolecule | Name: Circumsporozoite protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Plasmodium falciparum (malaria parasite P. falciparum) Plasmodium falciparum (malaria parasite P. falciparum) |
| Molecular weight | Theoretical: 39.986016 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: FQEYQCYGSS SNTRVLNELN YDNAGTNLYN ELEMNYYGKQ ENWYSLKKNS RSLGENDDGN NEDNEKLRKP KHKKLKQPAD GNPDPNANP NVDPNANPNV DPNANPNVDP NANPNANPNA NPNANPNANP NANPNANPNA NPNANPNANP NANPNANPNA N PNANPNAN ...String: FQEYQCYGSS SNTRVLNELN YDNAGTNLYN ELEMNYYGKQ ENWYSLKKNS RSLGENDDGN NEDNEKLRKP KHKKLKQPAD GNPDPNANP NVDPNANPNV DPNANPNVDP NANPNANPNA NPNANPNANP NANPNANPNA NPNANPNANP NANPNANPNA N PNANPNAN PNANPNANPN ANPNANPNAN PNANPNANPN ANPNANPNAN PNANPNANPN ANPNANPNAN PNANPNANPN AN PNANPNA NPNANPNKNN QGNGQGHNMP NDPNRNVDEN ANANSAVKNN NNEEPSDKHI KEYLNKIQNS LSTEWSPCSV TCG NGIQVR IKPGSANKPK DELDYANDIE KKICKMEKCS SVFNVVQSSP HHHHHH |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Homemade / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 4306 / Average electron dose: 45.24 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 1295064 |
|---|---|
| Initial angle assignment | Type: OTHER |
| Final 3D classification | Software - Name: cryoSPARC (ver. 2) |
| Final angle assignment | Type: NOT APPLICABLE |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2) / Number images used: 165747 |
 Movie
Movie Controller
Controller


 Z
Z Y
Y X
X