+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tetrahymena Polymerase alpha-Primase | ||||||||||||
 Map data Map data | full map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | CST /  polymerase / polymerase /  primase / primase /  REPLICATION REPLICATION | ||||||||||||
| Function / homology |  Function and homology information Function and homology information DNA primase activity / mitotic DNA replication initiation / DNA primase activity / mitotic DNA replication initiation /  DNA replication, synthesis of primer / DNA replication, synthesis of primer /  Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / 4 iron, 4 sulfur cluster binding / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / 4 iron, 4 sulfur cluster binding /  DNA replication / DNA replication /  DNA-directed DNA polymerase / DNA-directed DNA polymerase /  DNA-directed DNA polymerase activity / DNA-directed DNA polymerase activity /  nucleotide binding / nucleotide binding /  DNA binding ... DNA binding ... DNA primase activity / mitotic DNA replication initiation / DNA primase activity / mitotic DNA replication initiation /  DNA replication, synthesis of primer / DNA replication, synthesis of primer /  Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / 4 iron, 4 sulfur cluster binding / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / 4 iron, 4 sulfur cluster binding /  DNA replication / DNA replication /  DNA-directed DNA polymerase / DNA-directed DNA polymerase /  DNA-directed DNA polymerase activity / DNA-directed DNA polymerase activity /  nucleotide binding / nucleotide binding /  DNA binding / DNA binding /  metal ion binding / metal ion binding /  nucleus nucleusSimilarity search - Function | ||||||||||||
| Biological species |   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) | ||||||||||||
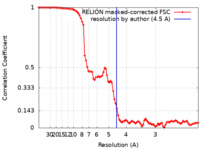
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.5 Å cryo EM / Resolution: 4.5 Å | ||||||||||||
 Authors Authors | He Y / Song H / Chan H / Wang Y / Liu B / Susac L / Zhou ZH / Feigon J | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structure of Tetrahymena telomerase-bound CST with polymerase α-primase. Authors: Yao He / He Song / Henry Chan / Baocheng Liu / Yaqiang Wang / Lukas Sušac / Z Hong Zhou / Juli Feigon /  Abstract: Telomeres are the physical ends of linear chromosomes. They are composed of short repeating sequences (such as TTGGGG in the G-strand for Tetrahymena thermophila) of double-stranded DNA with a single- ...Telomeres are the physical ends of linear chromosomes. They are composed of short repeating sequences (such as TTGGGG in the G-strand for Tetrahymena thermophila) of double-stranded DNA with a single-strand 3' overhang of the G-strand and, in humans, the six shelterin proteins: TPP1, POT1, TRF1, TRF2, RAP1 and TIN2. TPP1 and POT1 associate with the 3' overhang, with POT1 binding the G-strand and TPP1 (in complex with TIN2) recruiting telomerase via interaction with telomerase reverse transcriptase (TERT). The telomere DNA ends are replicated and maintained by telomerase, for the G-strand, and subsequently DNA polymerase α-primase (PolαPrim), for the C-strand. PolαPrim activity is stimulated by the heterotrimeric complex CTC1-STN1-TEN1 (CST), but the structural basis of the recruitment of PolαPrim and CST to telomere ends remains unknown. Here we report cryo-electron microscopy (cryo-EM) structures of Tetrahymena CST in the context of the telomerase holoenzyme, in both the absence and the presence of PolαPrim, and of PolαPrim alone. Tetrahymena Ctc1 binds telomerase subunit p50, a TPP1 orthologue, on a flexible Ctc1 binding motif revealed by cryo-EM and NMR spectroscopy. The PolαPrim polymerase subunit POLA1 binds Ctc1 and Stn1, and its interface with Ctc1 forms an entry port for G-strand DNA to the POLA1 active site. We thus provide a snapshot of four key components that are required for telomeric DNA synthesis in a single active complex-telomerase-core ribonucleoprotein, p50, CST and PolαPrim-that provides insights into the recruitment of CST and PolαPrim and the handoff between G-strand and C-strand synthesis. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26867.map.gz emd_26867.map.gz | 49.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26867-v30.xml emd-26867-v30.xml emd-26867.xml emd-26867.xml | 23.9 KB 23.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26867_fsc.xml emd_26867_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_26867.png emd_26867.png | 50.7 KB | ||
| Masks |  emd_26867_msk_1.map emd_26867_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26867.cif.gz emd-26867.cif.gz | 7.4 KB | ||
| Others |  emd_26867_additional_1.map.gz emd_26867_additional_1.map.gz emd_26867_half_map_1.map.gz emd_26867_half_map_1.map.gz emd_26867_half_map_2.map.gz emd_26867_half_map_2.map.gz | 49.1 MB 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26867 http://ftp.pdbj.org/pub/emdb/structures/EMD-26867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26867 | HTTPS FTP |
-Related structure data
| Related structure data |  7uy8MC  7uy5C  7uy6C  7uy7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26867.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26867.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
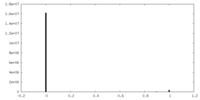
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26867_msk_1.map emd_26867_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
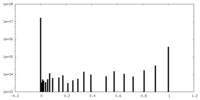
| Projections & Slices |
| ||||||||||||
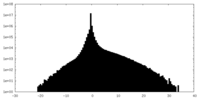
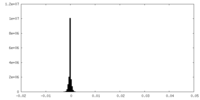
| Density Histograms |
-Additional map: composite map generated using two focused refined maps, sharpened
| File | emd_26867_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | composite map generated using two focused refined maps, sharpened | ||||||||||||
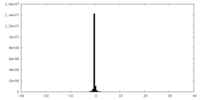
| Projections & Slices |
| ||||||||||||
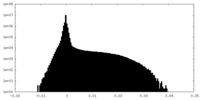
| Density Histograms |
-Half map: half1
| File | emd_26867_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
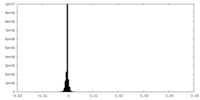
| Density Histograms |
-Half map: half2
| File | emd_26867_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
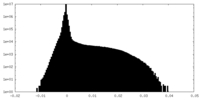
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrahymena PolaPrim
| Entire | Name: Tetrahymena PolaPrim |
|---|---|
| Components |
|
-Supramolecule #1: Tetrahymena PolaPrim
| Supramolecule | Name: Tetrahymena PolaPrim / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
-Macromolecule #1: DNA polymerase alpha subunit B
| Macromolecule | Name: DNA polymerase alpha subunit B / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
| Molecular weight | Theoretical: 68.773758 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MEEFEKILEE IDSANLSQNA VQQLKQLIIS NGILDDEQLM EKWLFQLEAI IYNSKQKFLE AQLMDFKKNV LNQDIKKPQN DNFKKIKES EVQKNIDIIK GDADLFTKAS SAKKQTSTQQ ANNAAIDCEE QLRQQLGKES MLDPKRFSYN SNLPQVNFEK N QHQNSNNM ...String: MEEFEKILEE IDSANLSQNA VQQLKQLIIS NGILDDEQLM EKWLFQLEAI IYNSKQKFLE AQLMDFKKNV LNQDIKKPQN DNFKKIKES EVQKNIDIIK GDADLFTKAS SAKKQTSTQQ ANNAAIDCEE QLRQQLGKES MLDPKRFSYN SNLPQVNFEK N QHQNSNNM TIVDLFEQYD PNKIPNYLTN NIDKIRKHLE QRILTFKIQN YSQFIVNGEQ LINDVSSYSK TEEVKMVGRL IS TENGFLN TTNLRIELNR NQYFDLVFEN DFDFGNNVLF PNQIVMVKGI INEKEELVVS ELITDHIKEI TDEEHPKIDN LNQ DESQLI MVAAGPFSTV MSTQYTSFDN ILHVAKTKKV STLILLGPFL DIKNEILKDG SIIINSKEYT FEQLQNSLFE KAVK ELEGK TQIIIVPSTR DILSTDSIPQ MSLEIKVSEH KNSIHSYSNP AYIDIDKLRV YIANSDVGMI TLQNSLLDKK IPYMQ QSRL TFKALMNQQN LYSIYPMRNP QDSQQILETV KLPNINDFDI PFDYQLEQYP HIIISPSSLP KFATKIYNTV VINPNY VIR DGSQAGNFAI ITLFKDSDIP IHERTRVDLY CL UniProtKB:  DNA polymerase alpha subunit B DNA polymerase alpha subunit B |
-Macromolecule #2: DNA polymerase
| Macromolecule | Name: DNA polymerase / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number:  DNA-directed DNA polymerase DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
| Molecular weight | Theoretical: 161.986984 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MSDKLTRLER LNKEVKKQNK LKQHSKNNRF DDDMDIEAYE DDEQIEEDDF IDDTQEDKKY KKKYREIEDE FDQEIEEEEE LNKKKKTKN TILNYTNTTA VTNNKKKAIS KQIPDIDIEE IMKLTERKKK IEQEEAQLLQ EEQELLEQEK REEEEKKRIS Q EAKSILRE ...String: MSDKLTRLER LNKEVKKQNK LKQHSKNNRF DDDMDIEAYE DDEQIEEDDF IDDTQEDKKY KKKYREIEDE FDQEIEEEEE LNKKKKTKN TILNYTNTTA VTNNKKKAIS KQIPDIDIEE IMKLTERKKK IEQEEAQLLQ EEQELLEQEK REEEEKKRIS Q EAKSILRE DCASSKTANS SKGKVDQNIL NAINRDFSDD SNTVDSISEF QKLKSLAQKA NLANESLKQS KVSNTEINLT NL SISQVKK INDYKNEDGS VDAYLYDYFY DAQVKPDKIY AFAKVQNKQT NAFDTCVIQI DTIIRNLFFY PSSDTVTEQQ IKN EIAELL KKEQTSRKNV EFLGAFVDKN YAFELPIPRG KSRWYQVVMS YEYEVISPDT KGQYFSYCVG STYSALETFL ITKK ITGPS WVRFQNVKDT TSCITNRKLE FRVDYTNQSN IQVLQKQLPT PPLSVVCISL KTSQQIVLSQ KKKEYKKEIF NLNMK YHEG INIDNSNKDE LNQFKSISFI THIDPTKKQD SITKKGTLPE TTKFCLNELN LLEQFLVHFN EIDPDIVVAH DLYSTV FEI ILTRIREKGI RKWNLLSKLI NIGSSDIPKY GSSTFKTKMA MKGRLLVDTL LSSQEFVNCV EYTLEALAQK LFKIEIP RI DAKAYQQKFA TYKLLNSLVD DTYQDIDYAL RIMYHLQIVP LTKQLTSICG NIWMGSLQNQ RAERNEMLLL HKFNQLNY V YPDNFKNLPE SYKKKHKNAQ IRKQYEEDED QAQGNKNPKK KENKYKGGQV FEPEKGLYNE YIVLLDFNSL YPSIIQEFN VCFTTCVRDP IPLEMQMAPF LGNKKAAIQY SKNQNTKENK MQDEDEEDNE NEQIVQTHDV LPTIEVIKGI APLPSILQYL VEQRKVVKN QIKGQKDPQV IETLDIKQKA FKLVANSMYG CLGFSSSRFY AMPLASFITA KGRHILFDSK KIVEDMGYSV I YGDTDSLM IKPGTNEFLE AVKTGLSIKI KVNSKYKKLQ LDIDGVFKNM LLLKKKKYAT LKVANWEEVK NTNAPEKLEK EI KGIDVVR RDWCQLSRDA GNKILEIILE SKSSENMLDD IKKYLIQLND DINQKNIKNS NYYITKRLTK RVDQYGEKNL PHV AVAQRS IQEKGIDPQT YVNQIISYII CKNEQSSRLV DKAYSPQEFI TQSKSLEIDL QYYKRFQLFE PIKRMLEVIE GINL QEIAS ILEVHYSVQH VSQNNELNAE NVLNLKSKRN QFLTSIPRVL VDCKKCDQTF LFLGILEENA DAASILKCKC GNDIY IQLK NKIALVVKEL IRNFEENAIQ IDNEEFEYTH QISLVGKAKQ QKMSSFTLNQ KLLSIQAMFD ITKEEQENTQ KVTIEK IKT IKKTLDDLLS KSQYNNLNLS NIFTSFGLLK UniProtKB:  DNA polymerase DNA polymerase |
-Macromolecule #3: Eukaryotic-type DNA primase, large subunit
| Macromolecule | Name: Eukaryotic-type DNA primase, large subunit / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
| Molecular weight | Theoretical: 64.386645 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MINNKQIQKI QQISKFQLKE YIEMQMQQHS LNKQIKEEER NYILLENAID NSNIQGFQTI QEIVQLAMQR MKMIKEIIQE EPRFNLIDP LSKPNYNRIR VANKFITVAI PKSDPEYELK NKQQYAKEVL SHFLCKMAYA FYAEPETWLE FARAEAFILM D KLKSGQHH ...String: MINNKQIQKI QQISKFQLKE YIEMQMQQHS LNKQIKEEER NYILLENAID NSNIQGFQTI QEIVQLAMQR MKMIKEIIQE EPRFNLIDP LSKPNYNRIR VANKFITVAI PKSDPEYELK NKQQYAKEVL SHFLCKMAYA FYAEPETWLE FARAEAFILM D KLKSGQHH ANFFSDENLK IKTISDELFK QAFPKIEATF SSIKIKKNDS EVEQINIKKD NFKMFKFIDH PSMLSNNDVV LH KGYIIIY KESTSKIVQN IFIERLLDEM RLLQLKFQNN GSKLDDDRLS FLKDLHKAEI FNDSTQFNST QIHHYELDRL AKR DMPACM TYLMYGLNQK LHLKHFGRLQ LGLFLKGAGL SLNEALTFWQ KKFSKTSADD FKKKYDYNIR HNYGKLGKQL DYTP MSCQK IIGFQPLKDE FHGCPYKTME SQQLKDFLKL SYNITDEQFV QVNIFKNQKQ YQLACKEVFK VLNDTRDKTK EQFYP YFDK VGNHPNAYFE QSLRMHEPQR FQKQDEEKKQ NKQNRNQNFS ANKQSTNKNN QMDLEF UniProtKB: Eukaryotic-type DNA primase, large subunit |
-Macromolecule #4: DNA primase
| Macromolecule | Name: DNA primase / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO EC number:  Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) |
| Molecular weight | Theoretical: 47.172855 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MEIETVEVAP QQEEEFKLDY DLLEEYYRSY FPVSQMVQWL SYPQDGDDTY FTRREFSFTL QNEVYLRYQM YNSEREFKNA LLQKVPEKI DIGAVYDRPG KKGDDIKAKE KEFVIDIDMT DYDHIRTCCS KAKICEKCWK FMRVACDLIS KSLDEDFGFQ H VLWVYSGR ...String: MEIETVEVAP QQEEEFKLDY DLLEEYYRSY FPVSQMVQWL SYPQDGDDTY FTRREFSFTL QNEVYLRYQM YNSEREFKNA LLQKVPEKI DIGAVYDRPG KKGDDIKAKE KEFVIDIDMT DYDHIRTCCS KAKICEKCWK FMRVACDLIS KSLDEDFGFQ H VLWVYSGR RGIHAWVCDK EIRKANDYTR ASIIDYLNIL VDNSIGSSYV KPSLLKMEKS HLIERNAMKQ LNQKNDDLKA EK ESEKVFV EIVLREQNLF MKKPEIILEF LAKRSENLSK EVEKEWKTLK TSEQRYEALK ELVSSEDKKK THYLLEELRI WLL YPRLDV NVSKSTNHLL KSPFCIHPKT GNVCVPFTTE EISTFDPFSV PNISTLTTEE GSSKMKNSLK IFNKFLENLK KDV UniProtKB:  DNA primase DNA primase |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z
Z Y
Y X
X