+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tetrahymena telomerase with CST | ||||||||||||
 Map data Map data | Tetrahymena telomerase with CST | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  telomerase / RNP / CST / telomerase / RNP / CST /  REPLICATION REPLICATION | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationtelomerase catalytic core complex assembly / telomerase RNA stabilization / telomerase RNA reverse transcriptase activity / DNA replication factor A complex / single-stranded telomeric DNA binding /  telomerase RNA binding / telomerase RNA binding /  telomerase holoenzyme complex / telomeric DNA binding / telomere maintenance via telomerase / telomerase holoenzyme complex / telomeric DNA binding / telomere maintenance via telomerase /  RNA-directed DNA polymerase ...telomerase catalytic core complex assembly / telomerase RNA stabilization / telomerase RNA reverse transcriptase activity / DNA replication factor A complex / single-stranded telomeric DNA binding / RNA-directed DNA polymerase ...telomerase catalytic core complex assembly / telomerase RNA stabilization / telomerase RNA reverse transcriptase activity / DNA replication factor A complex / single-stranded telomeric DNA binding /  telomerase RNA binding / telomerase RNA binding /  telomerase holoenzyme complex / telomeric DNA binding / telomere maintenance via telomerase / telomerase holoenzyme complex / telomeric DNA binding / telomere maintenance via telomerase /  RNA-directed DNA polymerase / DNA recombination / RNA-directed DNA polymerase / DNA recombination /  DNA replication / DNA replication /  chromosome, telomeric region / chromosome, telomeric region /  DNA repair / DNA repair /  DNA binding / zinc ion binding / DNA binding / zinc ion binding /  metal ion binding metal ion bindingSimilarity search - Function | ||||||||||||
| Biological species |   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) | ||||||||||||
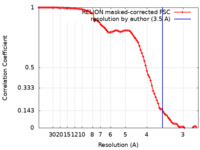
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | He Y / Song H / Chan H / Wang Y / Liu B / Susac L / Zhou ZH / Feigon J | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structure of Tetrahymena telomerase-bound CST with polymerase α-primase. Authors: Yao He / He Song / Henry Chan / Baocheng Liu / Yaqiang Wang / Lukas Sušac / Z Hong Zhou / Juli Feigon /  Abstract: Telomeres are the physical ends of linear chromosomes. They are composed of short repeating sequences (such as TTGGGG in the G-strand for Tetrahymena thermophila) of double-stranded DNA with a single- ...Telomeres are the physical ends of linear chromosomes. They are composed of short repeating sequences (such as TTGGGG in the G-strand for Tetrahymena thermophila) of double-stranded DNA with a single-strand 3' overhang of the G-strand and, in humans, the six shelterin proteins: TPP1, POT1, TRF1, TRF2, RAP1 and TIN2. TPP1 and POT1 associate with the 3' overhang, with POT1 binding the G-strand and TPP1 (in complex with TIN2) recruiting telomerase via interaction with telomerase reverse transcriptase (TERT). The telomere DNA ends are replicated and maintained by telomerase, for the G-strand, and subsequently DNA polymerase α-primase (PolαPrim), for the C-strand. PolαPrim activity is stimulated by the heterotrimeric complex CTC1-STN1-TEN1 (CST), but the structural basis of the recruitment of PolαPrim and CST to telomere ends remains unknown. Here we report cryo-electron microscopy (cryo-EM) structures of Tetrahymena CST in the context of the telomerase holoenzyme, in both the absence and the presence of PolαPrim, and of PolαPrim alone. Tetrahymena Ctc1 binds telomerase subunit p50, a TPP1 orthologue, on a flexible Ctc1 binding motif revealed by cryo-EM and NMR spectroscopy. The PolαPrim polymerase subunit POLA1 binds Ctc1 and Stn1, and its interface with Ctc1 forms an entry port for G-strand DNA to the POLA1 active site. We thus provide a snapshot of four key components that are required for telomeric DNA synthesis in a single active complex-telomerase-core ribonucleoprotein, p50, CST and PolαPrim-that provides insights into the recruitment of CST and PolαPrim and the handoff between G-strand and C-strand synthesis. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26863.map.gz emd_26863.map.gz | 5.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26863-v30.xml emd-26863-v30.xml emd-26863.xml emd-26863.xml | 28.6 KB 28.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26863_fsc.xml emd_26863_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_26863.png emd_26863.png | 98.2 KB | ||
| Masks |  emd_26863_msk_1.map emd_26863_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26863.cif.gz emd-26863.cif.gz | 8.7 KB | ||
| Others |  emd_26863_half_map_1.map.gz emd_26863_half_map_1.map.gz emd_26863_half_map_2.map.gz emd_26863_half_map_2.map.gz | 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26863 http://ftp.pdbj.org/pub/emdb/structures/EMD-26863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26863 | HTTPS FTP |
-Related structure data
| Related structure data |  7uy5MC  7uy6C  7uy7C  7uy8C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26863.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26863.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tetrahymena telomerase with CST | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26863_msk_1.map emd_26863_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_26863_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_26863_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Tetrahymena telomerase holoenzyme
+Supramolecule #1: Tetrahymena telomerase holoenzyme
+Macromolecule #1: Telomerase La-related protein p65
+Macromolecule #2: Telomerase reverse transcriptase
+Macromolecule #3: Telomerase holoenzyme Teb1 subunit
+Macromolecule #4: Telomerase holoenzyme Teb2 subunit
+Macromolecule #5: Telomerase holoenzyme Teb3 subunit
+Macromolecule #6: Telomerase associated protein p50
+Macromolecule #9: Telomerase-associated protein of 75 kDa
+Macromolecule #10: Telomerase-associated protein of 19 kDa
+Macromolecule #11: Telomerase-associated protein of 45 kDa
+Macromolecule #7: Telomerase RNA
+Macromolecule #8: Telomere DNA
+Macromolecule #12: ZINC ION
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 48.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X