+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TnsBctd-TnsC-TniQ complex | |||||||||
 Map data Map data | LocSpiral processed map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  [Scytonema hofmanni] UTEX 2349 (bacteria) / synthetic construct (others) [Scytonema hofmanni] UTEX 2349 (bacteria) / synthetic construct (others) | |||||||||
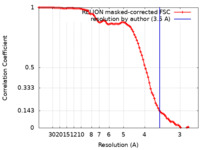
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Park J / Tsai AWT / Kellogg EH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structures of the holo CRISPR RNA-guided transposon integration complex. Authors: Jung-Un Park / Amy Wei-Lun Tsai / Alexandrea N Rizo / Vinh H Truong / Tristan X Wellner / Richard D Schargel / Elizabeth H Kellogg /  Abstract: CRISPR-associated transposons (CAST) are programmable mobile genetic elements that insert large DNA cargos using an RNA-guided mechanism. CAST elements contain multiple conserved proteins: a CRISPR ...CRISPR-associated transposons (CAST) are programmable mobile genetic elements that insert large DNA cargos using an RNA-guided mechanism. CAST elements contain multiple conserved proteins: a CRISPR effector (Cas12k or Cascade), a AAA+ regulator (TnsC), a transposase (TnsA-TnsB) and a target-site-associated factor (TniQ). These components are thought to cooperatively integrate DNA via formation of a multisubunit transposition integration complex (transpososome). Here we reconstituted the approximately 1 MDa type V-K CAST transpososome from Scytonema hofmannii (ShCAST) and determined its structure using single-particle cryo-electon microscopy. The architecture of this transpososome reveals modular association between the components. Cas12k forms a complex with ribosomal subunit S15 and TniQ, stabilizing formation of a full R-loop. TnsC has dedicated interaction interfaces with TniQ and TnsB. Of note, we observe TnsC-TnsB interactions at the C-terminal face of TnsC, which contribute to the stimulation of ATPase activity. Although the TnsC oligomeric assembly deviates slightly from the helical configuration found in isolation, the TnsC-bound target DNA conformation differs markedly in the transpososome. As a consequence, TnsC makes new protein-DNA interactions throughout the transpososome that are important for transposition activity. Finally, we identify two distinct transpososome populations that differ in their DNA contacts near TniQ. This suggests that associations with the CRISPR effector can be flexible. This ShCAST transpososome structure enhances our understanding of CAST transposition systems and suggests ways to improve CAST transposition for precision genome-editing applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25453.map.gz emd_25453.map.gz | 5.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25453-v30.xml emd-25453-v30.xml emd-25453.xml emd-25453.xml | 26 KB 26 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25453_fsc.xml emd_25453_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_25453.png emd_25453.png | 82.7 KB | ||
| Masks |  emd_25453_msk_1.map emd_25453_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_25453_additional_1.map.gz emd_25453_additional_1.map.gz emd_25453_additional_2.map.gz emd_25453_additional_2.map.gz emd_25453_half_map_1.map.gz emd_25453_half_map_1.map.gz emd_25453_half_map_2.map.gz emd_25453_half_map_2.map.gz | 49.5 MB 5.6 MB 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25453 http://ftp.pdbj.org/pub/emdb/structures/EMD-25453 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25453 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25453 | HTTPS FTP |
-Related structure data
| Related structure data |  7svuMC  8ea3C  8ea4C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25453.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25453.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocSpiral processed map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.33 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25453_msk_1.map emd_25453_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
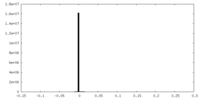
| Density Histograms |
-Additional map: Full map
| File | emd_25453_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
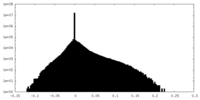
| Density Histograms |
-Additional map: uniform B-factor sharpened map
| File | emd_25453_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | uniform B-factor sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
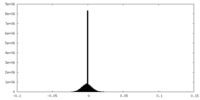
| Density Histograms |
-Half map: halfmap1
| File | emd_25453_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
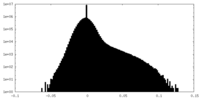
| Density Histograms |
-Half map: halfmap 2
| File | emd_25453_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ATP-bound TnsBctd-TnsC-TniQ complex from ShCAST element
| Entire | Name: ATP-bound TnsBctd-TnsC-TniQ complex from ShCAST element |
|---|---|
| Components |
|
-Supramolecule #1: ATP-bound TnsBctd-TnsC-TniQ complex from ShCAST element
| Supramolecule | Name: ATP-bound TnsBctd-TnsC-TniQ complex from ShCAST element type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  [Scytonema hofmanni] UTEX 2349 (bacteria) [Scytonema hofmanni] UTEX 2349 (bacteria) |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: DNA (28-MER)
| Macromolecule | Name: DNA (28-MER) / type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.472441 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) |
-Macromolecule #2: DNA (29-MER)
| Macromolecule | Name: DNA (29-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 9.03804 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) |
-Macromolecule #3: TnsC
| Macromolecule | Name: TnsC / type: protein_or_peptide / ID: 3 / Number of copies: 11 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  [Scytonema hofmanni] UTEX 2349 (bacteria) [Scytonema hofmanni] UTEX 2349 (bacteria) |
| Molecular weight | Theoretical: 31.444617 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MTEAQAIAKQ LGGVKPDDEW LQAEIARLKG KSIVPLQQVK TLHDWLDGKR KARKSCRVVG ESRTGKTVAC DAYRYRHKPQ QEAGRPPTV PVVYIRPHQK CGPKDLFKKI TEYLKYRVTK GTVSDFRDRT IEVLKGCGVE MLIIDEADRL KPETFADVRD I AEDLGIAV ...String: MTEAQAIAKQ LGGVKPDDEW LQAEIARLKG KSIVPLQQVK TLHDWLDGKR KARKSCRVVG ESRTGKTVAC DAYRYRHKPQ QEAGRPPTV PVVYIRPHQK CGPKDLFKKI TEYLKYRVTK GTVSDFRDRT IEVLKGCGVE MLIIDEADRL KPETFADVRD I AEDLGIAV VLVGTDRLDA VIKRDEQVLE RFRAHLRFGK LSGEDFKNTV EMWEQMVLKL PVSSNLKSKE MLRILTSATE GY IGRLDEI LREAAIRSLS RGLKKIDKAV LQEVAKEYK |
-Macromolecule #4: TnsB-CTD
| Macromolecule | Name: TnsB-CTD / type: protein_or_peptide / ID: 4 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  [Scytonema hofmanni] UTEX 2349 (bacteria) [Scytonema hofmanni] UTEX 2349 (bacteria) |
| Molecular weight | Theoretical: 1.97711 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: IEVWDYEQLR EEYGF |
-Macromolecule #5: TniQ
| Macromolecule | Name: TniQ / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  [Scytonema hofmanni] UTEX 2349 (bacteria) [Scytonema hofmanni] UTEX 2349 (bacteria) |
| Molecular weight | Theoretical: 19.01124 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MIEAPDVKPW LFLIKPYEGE SLSHFLGRFR RANHLSASGL GTLAGIGAIV ARWERFHFNP RPSQQELEAI ASVVEVDAQR LAQMLPPAG VGMQHEPIRL CGACYAESPC HRIEWQYKSV WKCDRHQLKI LAKCPNCQAP FKMPALWEDG CCHRCRMPFA E MAKLQKV |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 11 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 11 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Support film - Material: GOLD / Support film - topology: HOLEY | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7svu: |
 Movie
Movie Controller
Controller





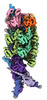




 Z
Z Y
Y X
X