[English] 日本語
 Yorodumi
Yorodumi- EMDB-2484: Pre-fusion structure of trimeric HIV-1 envelope glycoprotein dete... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2484 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
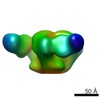
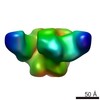
| Title | Pre-fusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy | |||||||||
 Map data Map data | Structure of HIV-1 gp140 trimer bound to VRC03 Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 envelope glycoprotein / SOSIP gp140 trimer / HIV-1 Env / pre-fusion state | |||||||||
| Biological species |    Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 6.0 Å cryo EM / Resolution: 6.0 Å | |||||||||
 Authors Authors | Bartesaghi A / Merk A / Borgnia MJ / Milne JLS / Subramaniam S | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2013 Journal: Nat Struct Mol Biol / Year: 2013Title: Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Authors: Alberto Bartesaghi / Alan Merk / Mario J Borgnia / Jacqueline L S Milne / Sriram Subramaniam /  Abstract: The activation of trimeric HIV-1 envelope glycoprotein (Env) by its binding to the cell-surface receptor CD4 and co-receptors (CCR5 or CXCR4) represents the first of a series of events that lead to ...The activation of trimeric HIV-1 envelope glycoprotein (Env) by its binding to the cell-surface receptor CD4 and co-receptors (CCR5 or CXCR4) represents the first of a series of events that lead to fusion between viral and target-cell membranes. Here, we present the cryo-EM structure, at subnanometer resolution (~6 Å at 0.143 FSC), of the 'closed', prefusion state of trimeric HIV-1 Env complexed to the broadly neutralizing antibody VRC03. We show that three gp41 helices at the core of the trimer serve as an anchor around which the rest of Env is reorganized upon activation to the 'open' quaternary conformation. The architecture of trimeric HIV-1 Env in the prefusion state and in the activated intermediate state resembles the corresponding states of influenza hemagglutinin trimers, thus providing direct evidence for the similarity in entry mechanisms used by HIV-1, influenza and related enveloped viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2484.map.gz emd_2484.map.gz | 55.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2484-v30.xml emd-2484-v30.xml emd-2484.xml emd-2484.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2484_square.png EMD-2484_square.png | 290.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2484 http://ftp.pdbj.org/pub/emdb/structures/EMD-2484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2484 | HTTPS FTP |
-Related structure data
| Related structure data |  4cc8MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10004 (Title: Pre-fusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy EMPIAR-10004 (Title: Pre-fusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopyData size: 117.8 Data #1: HIV-1 envelope glycoprotein micrographs [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2484.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2484.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of HIV-1 gp140 trimer bound to VRC03 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of KNH1144 SOSIP gp140 trimer with VRC03 Fab.
| Entire | Name: Complex of KNH1144 SOSIP gp140 trimer with VRC03 Fab. |
|---|---|
| Components |
|
-Supramolecule #1000: Complex of KNH1144 SOSIP gp140 trimer with VRC03 Fab.
| Supramolecule | Name: Complex of KNH1144 SOSIP gp140 trimer with VRC03 Fab. / type: sample / ID: 1000 Oligomeric state: Tetramer (gp41, gp120, Fab Heavy, Fab Light) Number unique components: 4 |
|---|---|
| Molecular weight | Theoretical: 670 KDa |
-Macromolecule #1: gp120
| Macromolecule | Name: gp120 / type: protein_or_peptide / ID: 1 / Name.synonym: Envelope glycoprotein / Number of copies: 3 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 / Strain: HIV-1 isolate 00KE_KNH1144 / synonym: HIV-1 Human immunodeficiency virus 1 / Strain: HIV-1 isolate 00KE_KNH1144 / synonym: HIV-1 |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant plasmid: SOSIP-PPI4 and furin-pcDNA3.1 Homo sapiens (human) / Recombinant plasmid: SOSIP-PPI4 and furin-pcDNA3.1 |
-Macromolecule #2: gp41
| Macromolecule | Name: gp41 / type: protein_or_peptide / ID: 2 / Name.synonym: Envelope Glycoprotein Details: Complete ectodomain of HIV-1 Env from the Clade A strain KNH1144 inlcuding residues in the membrane proximal external region with the following residue substitutions A662E, S668N, and S676T Number of copies: 3 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 / Strain: HIV-1 isolate 00KE_KNH1144 / synonym: HIV-1 Human immunodeficiency virus 1 / Strain: HIV-1 isolate 00KE_KNH1144 / synonym: HIV-1 |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant plasmid: SOSIP-PPI4 and furin-pcDNA3.1 Homo sapiens (human) / Recombinant plasmid: SOSIP-PPI4 and furin-pcDNA3.1 |
-Macromolecule #3: Monoclonal antibody VRC03 Fab Heavy chain
| Macromolecule | Name: Monoclonal antibody VRC03 Fab Heavy chain / type: protein_or_peptide / ID: 3 / Name.synonym: VRC03 Fab / Details: Fab fragment / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK 293 Homo sapiens (human) / Recombinant cell: HEK 293 |
-Macromolecule #4: Monoclonal antibody VRC03 Fab Light chain
| Macromolecule | Name: Monoclonal antibody VRC03 Fab Light chain / type: protein_or_peptide / ID: 4 / Name.synonym: VRC03 Fab / Details: Fab fragment / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK 293 Homo sapiens (human) / Recombinant cell: HEK 293 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.65 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: TNE Buffer (10 mM Tris, 150 mM NaCl, 1 mM EDTA) |
| Grid | Details: Protochips C-flat R 2/2, plasma cleaned |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV Method: blot for 6 seconds, blot offset of -2, plunge into an ethane slurry cooled by liquid nitrogen |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 80 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 75000 Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Date | Aug 9, 2011 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 4713 / Average electron dose: 10 e/Å2 / Bits/pixel: 16 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final two d classification | Number classes: 500 |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 6.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: EMAN, IMAGIC, FREALIGN / Number images used: 88125 ) / Resolution.type: BY AUTHOR / Resolution: 6.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: EMAN, IMAGIC, FREALIGN / Number images used: 88125 |
| Details | Particles were selected manually, classified in 2D using IMAGIC and 3D refinement was done with FREALIGN. |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: G / Chain - #1 - Chain ID: H / Chain - #2 - Chain ID: L |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | 3SE8, Chain G and chains H, L fitted as two separate rigid bodies into map. 3HMG helix fitted by hand into map. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-4cc8: |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - Chain ID: B |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid body. Automated fitting procedures. Only residues 92-122 of chain B were fitted. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-4cc8: |
 Movie
Movie Controller
Controller