[English] 日本語
 Yorodumi
Yorodumi- EMDB-24680: Peptide-19 bound to the Glucagon-Like Peptide-1 Receptor (GLP-1R) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24680 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Peptide-19 bound to the Glucagon-Like Peptide-1 Receptor (GLP-1R) | ||||||||||||||||||||||||
 Map data Map data | The sharpened cryoSPARC consensus map at 2.14A. | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information glucagon-like peptide 1 receptor activity / glucagon-like peptide 1 receptor activity /  glucagon receptor activity / hormone secretion / positive regulation of blood pressure / glucagon receptor activity / hormone secretion / positive regulation of blood pressure /  post-translational protein targeting to membrane, translocation / post-translational protein targeting to membrane, translocation /  regulation of heart contraction / response to psychosocial stress / PKA activation in glucagon signalling / regulation of heart contraction / response to psychosocial stress / PKA activation in glucagon signalling /  peptide hormone binding / cAMP-mediated signaling ... peptide hormone binding / cAMP-mediated signaling ... glucagon-like peptide 1 receptor activity / glucagon-like peptide 1 receptor activity /  glucagon receptor activity / hormone secretion / positive regulation of blood pressure / glucagon receptor activity / hormone secretion / positive regulation of blood pressure /  post-translational protein targeting to membrane, translocation / post-translational protein targeting to membrane, translocation /  regulation of heart contraction / response to psychosocial stress / PKA activation in glucagon signalling / regulation of heart contraction / response to psychosocial stress / PKA activation in glucagon signalling /  peptide hormone binding / cAMP-mediated signaling / hair follicle placode formation / developmental growth / peptide hormone binding / cAMP-mediated signaling / hair follicle placode formation / developmental growth /  D1 dopamine receptor binding / D1 dopamine receptor binding /  intracellular transport / Hedgehog 'off' state / positive regulation of cAMP-mediated signaling / adenylate cyclase-activating adrenergic receptor signaling pathway / activation of adenylate cyclase activity / adenylate cyclase activator activity / negative regulation of blood pressure / trans-Golgi network membrane / Olfactory Signaling Pathway / G-protein beta/gamma-subunit complex binding / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / intracellular transport / Hedgehog 'off' state / positive regulation of cAMP-mediated signaling / adenylate cyclase-activating adrenergic receptor signaling pathway / activation of adenylate cyclase activity / adenylate cyclase activator activity / negative regulation of blood pressure / trans-Golgi network membrane / Olfactory Signaling Pathway / G-protein beta/gamma-subunit complex binding / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor /  bone development / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / bone development / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion /  platelet aggregation / platelet aggregation /  cognition / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / positive regulation of GTPase activity / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / Inactivation, recovery and regulation of the phototransduction cascade / cognition / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / positive regulation of GTPase activity / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / Inactivation, recovery and regulation of the phototransduction cascade /  heterotrimeric G-protein complex / G alpha (12/13) signalling events / heterotrimeric G-protein complex / G alpha (12/13) signalling events /  extracellular vesicle / signaling receptor complex adaptor activity / sensory perception of smell / transmembrane signaling receptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / extracellular vesicle / signaling receptor complex adaptor activity / sensory perception of smell / transmembrane signaling receptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye /  GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cold-induced thermogenesis / G alpha (i) signalling events / positive regulation of cytosolic calcium ion concentration / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / learning or memory / cell surface receptor signaling pathway / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cold-induced thermogenesis / G alpha (i) signalling events / positive regulation of cytosolic calcium ion concentration / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / learning or memory / cell surface receptor signaling pathway / G protein-coupled receptor signaling pathway / lysosomal membrane /  GTPase activity / GTPase activity /  synapse / protein-containing complex binding / GTP binding / synapse / protein-containing complex binding / GTP binding /  signal transduction / extracellular exosome / signal transduction / extracellular exosome /  membrane / membrane /  metal ion binding / metal ion binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||||||||||||||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Lama glama (llama) / synthetic construct (others) Lama glama (llama) / synthetic construct (others) | ||||||||||||||||||||||||
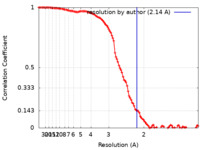
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.14 Å cryo EM / Resolution: 2.14 Å | ||||||||||||||||||||||||
 Authors Authors | Johnson RM / Danev R / Sexton PM / Wootten D | ||||||||||||||||||||||||
| Funding support |  Japan, 7 items Japan, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Biochem Biophys Res Commun / Year: 2021 Journal: Biochem Biophys Res Commun / Year: 2021Title: Cryo-EM structure of the dual incretin receptor agonist, peptide-19, in complex with the glucagon-like peptide-1 receptor. Authors: Rachel M Johnson / Xin Zhang / Sarah J Piper / Theodore J Nettleton / Teresa H Vandekolk / Christopher J Langmead / Radostin Danev / Patrick M Sexton / Denise Wootten /   Abstract: Dual agonists that can activate both the glucagon-like peptide-1 receptor (GLP-1R) and the gastric inhibitory polypeptide receptor (GIPR) have demonstrated high efficacy for the treatment of ...Dual agonists that can activate both the glucagon-like peptide-1 receptor (GLP-1R) and the gastric inhibitory polypeptide receptor (GIPR) have demonstrated high efficacy for the treatment of metabolic disease. Peptide-19 is a prototypical dual agonist that has high potency at both GLP-1R and GIPR but has a distinct signalling profile relative to the native peptides at the cognate receptors. In this study, we solved the structure of peptide-19 bound to the GLP-1R in complex with Gs protein, and compared the structure and dynamics of this complex to that of published structures of GLP-1R:Gs in complex with other receptor agonists. Unlike other peptide-bound receptor complexes, peptide-19:GLP-1R:Gs demonstrated a more open binding pocket where transmembrane domain (TM) 6, TM7 and the interconnecting extracellular loop 3 (ECL3) were located away from the peptide, with no interactions between peptide-19 and TM6/ECL3. Analysis of conformational variance of the complex revealed that peptide-19 was highly dynamic and underwent binding and unbinding motions facilitated by the more open TM binding pocket. Both the consensus structure of the GLP-1R complex with peptide-19 and the dynamics of this complex were distinct from previously described GLP-1R structures providing unique insights into the mode of GLP-1R activation by this dual agonist. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24680.map.gz emd_24680.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24680-v30.xml emd-24680-v30.xml emd-24680.xml emd-24680.xml | 44.1 KB 44.1 KB | Display Display |  EMDB header EMDB header |
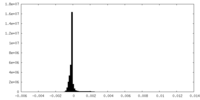
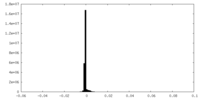
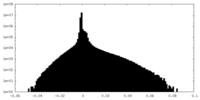
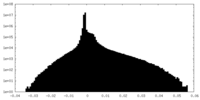
| FSC (resolution estimation) |  emd_24680_fsc.xml emd_24680_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_24680.png emd_24680.png | 41.4 KB | ||
| Masks |  emd_24680_msk_1.map emd_24680_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_24680_additional_1.map.gz emd_24680_additional_1.map.gz emd_24680_additional_10.map.gz emd_24680_additional_10.map.gz emd_24680_additional_2.map.gz emd_24680_additional_2.map.gz emd_24680_additional_3.map.gz emd_24680_additional_3.map.gz emd_24680_additional_4.map.gz emd_24680_additional_4.map.gz emd_24680_additional_5.map.gz emd_24680_additional_5.map.gz emd_24680_additional_6.map.gz emd_24680_additional_6.map.gz emd_24680_additional_7.map.gz emd_24680_additional_7.map.gz emd_24680_additional_8.map.gz emd_24680_additional_8.map.gz emd_24680_additional_9.map.gz emd_24680_additional_9.map.gz emd_24680_half_map_1.map.gz emd_24680_half_map_1.map.gz emd_24680_half_map_2.map.gz emd_24680_half_map_2.map.gz | 61.5 MB 84.4 MB 108.3 MB 7.2 MB 10.8 MB 111 MB 110.9 MB 82.6 MB 82.8 MB 79.9 MB 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24680 http://ftp.pdbj.org/pub/emdb/structures/EMD-24680 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24680 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24680 | HTTPS FTP |
-Related structure data
| Related structure data |  7rtbMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24680.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24680.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The sharpened cryoSPARC consensus map at 2.14A. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Mask #1
+Additional map: The unsharpened cryoSPARC consensus map.
+Additional map: The local resolution map for the ECD-focussed refinement...
+Additional map: Unsharpened receptor-only focussed refinement from RELION
+Additional map: Sharpened receptor-only focussed refinement from RELION
+Additional map: Sharpened consensus refinement generated in RELION
+Additional map: Unsharpened ECD-only focussed refinement from RELION
+Additional map: Unsharpened consensus refinement generated in RELION
+Additional map: The local resolution map for the receptor-focussed refinement...
+Additional map: The local resolution map for the consensus map generated using RELION
+Additional map: The local resolution map for the best-resolved ECD...
+Half map: Half map B from cryoSPARC consensus refinement.
+Half map: Half map A from cryoSPARC consensus refinement.
- Sample components
Sample components
-Entire : Peptide-19-GLP-1R-Gs complex
| Entire | Name: Peptide-19-GLP-1R-Gs complex |
|---|---|
| Components |
|
-Supramolecule #1: Peptide-19-GLP-1R-Gs complex
| Supramolecule | Name: Peptide-19-GLP-1R-Gs complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.683434 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQA DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ AALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENIRRVF NDCRDIIQRM HLRQYELL |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.285734 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW ...String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW DIETGQQTTT FTGHTGDVMS LSLAPDTRLF VSGACDASAK LWDVREGMCR QTFTGHESDI NAICFFPNGN AF ATGSDDA TCRLFDLRAD QELMTYSHDN IICGITSVSF SKSGRLLLAG YDDFNCNVWD ALKADRAGVL AGHDNRVSCL GVT DDGMAV ATGSWDSFLK IWN |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.375332 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: NTASIAQARK LVEQLKMEAN IDRIKVSKAA ADLMAYCEAH AKEDPLLTPV PASENPFR |
-Macromolecule #4: Nb35
| Macromolecule | Name: Nb35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Lama glama (llama) Lama glama (llama) |
| Molecular weight | Theoretical: 13.885439 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSS |
-Macromolecule #5: Glucagon-like peptide 1 receptor
| Macromolecule | Name: Glucagon-like peptide 1 receptor / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.668316 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDLEVLFQG PRPQGATVSL WETVQKWREY RRQCQRSLTE DPPPATDLFC NRTFDEYACW PDGEPGSFV NVSCPWYLPW ASSVPQGHVY RFCTAEGLWL QKDNSSLPWR DLSECEESKR GERSSPEEQL LFLYIIYTVG Y ALSFSALV ...String: MKTIIALSYI FCLVFADYKD DDDLEVLFQG PRPQGATVSL WETVQKWREY RRQCQRSLTE DPPPATDLFC NRTFDEYACW PDGEPGSFV NVSCPWYLPW ASSVPQGHVY RFCTAEGLWL QKDNSSLPWR DLSECEESKR GERSSPEEQL LFLYIIYTVG Y ALSFSALV IASAILLGFR HLHCTRNYIH LNLFASFILR ALSVFIKDAA LKWMYSTAAQ QHQWDGLLSY QDSLSCRLVF LL MQYCVAA NYYWLLVEGV YLYTLLAFSV FSEQWIFRLY VSIGWGVPLL FVVPWGIVKY LYEDEGCWTR NSNMNYWLII RLP ILFAIG VNFLIFVRVI CIVVSKLKAN LMCKTDIKCR LAKSTLTLIP LLGTHEVIFA FVMDEHARGT LRHIKLFTEL SFTS FQGLM VAILYCFVNN EVQLEFRKSW ERWRLEHLHI QRDSSMKPLK CPTSSLSSGA TAGSSMYTAT CQASCSPAGL EVLFQ GPHH HHHHHH |
-Macromolecule #6: Peptide-19
| Macromolecule | Name: Peptide-19 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.093503 KDa |
| Sequence | String: Y(AIB)EGTFTSDY SIYLDKQAA(AIB) EFVNWLLAGG PSAPPPSK |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal magnification: 130000 Bright-field microscopy / Cs: 2.7 mm / Nominal magnification: 130000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 15 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6858 / Average exposure time: 4.352 sec. / Average electron dose: 70.15 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
































 Z
Z Y
Y X
X