[English] 日本語
 Yorodumi
Yorodumi- EMDB-2427: Negative stain Electron Microscopy of BG505 SOSIP.664 gp140 in co... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2427 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
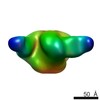
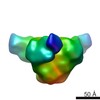
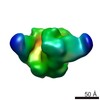
| Title | Negative stain Electron Microscopy of BG505 SOSIP.664 gp140 in complex with PGV04 | |||||||||
 Map data Map data | Bg505 SOSIP.664 with PGV04 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV antibody Env trimer / Bg505 SOSIP.664 / PGV04 | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / structural molecule activity / virion attachment to host cell / host cell plasma membrane / virion membrane / identical protein binding / viral envelope / structural molecule activity / virion attachment to host cell / host cell plasma membrane / virion membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |    Human immunodeficiency virus / Human immunodeficiency virus /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 23.0 Å negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Sanders RW / Derking R / Cupo A / Julien JP / Yasmeen A / de Val N / Kim HJ / Blattner C / Torrents A / Korzun J ...Sanders RW / Derking R / Cupo A / Julien JP / Yasmeen A / de Val N / Kim HJ / Blattner C / Torrents A / Korzun J / Golabek M / de los Reyes K / Ketas TJ / van Gils MJ / King CR / Wilson IA / Ward AB / Klasse PJ / Moore JP | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2013 Journal: PLoS Pathog / Year: 2013Title: A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. Authors: Rogier W Sanders / Ronald Derking / Albert Cupo / Jean-Philippe Julien / Anila Yasmeen / Natalia de Val / Helen J Kim / Claudia Blattner / Alba Torrents de la Peña / Jacob Korzun / Michael ...Authors: Rogier W Sanders / Ronald Derking / Albert Cupo / Jean-Philippe Julien / Anila Yasmeen / Natalia de Val / Helen J Kim / Claudia Blattner / Alba Torrents de la Peña / Jacob Korzun / Michael Golabek / Kevin de Los Reyes / Thomas J Ketas / Marit J van Gils / C Richter King / Ian A Wilson / Andrew B Ward / P J Klasse / John P Moore /  Abstract: A desirable but as yet unachieved property of a human immunodeficiency virus type 1 (HIV-1) vaccine candidate is the ability to induce broadly neutralizing antibodies (bNAbs). One approach to the ...A desirable but as yet unachieved property of a human immunodeficiency virus type 1 (HIV-1) vaccine candidate is the ability to induce broadly neutralizing antibodies (bNAbs). One approach to the problem is to create trimeric mimics of the native envelope glycoprotein (Env) spike that expose as many bNAb epitopes as possible, while occluding those for non-neutralizing antibodies (non-NAbs). Here, we describe the design and properties of soluble, cleaved SOSIP.664 gp140 trimers based on the subtype A transmitted/founder strain, BG505. These trimers are highly stable, more so even than the corresponding gp120 monomer, as judged by differential scanning calorimetry. They are also homogenous and closely resemble native virus spikes when visualized by negative stain electron microscopy (EM). We used several techniques, including ELISA and surface plasmon resonance (SPR), to determine the relationship between the ability of monoclonal antibodies (MAbs) to bind the soluble trimers and neutralize the corresponding virus. In general, the concordance was excellent, in that virtually all bNAbs against multiple neutralizing epitopes on HIV-1 Env were highly reactive with the BG505 SOSIP.664 gp140 trimers, including quaternary epitopes (CH01, PG9, PG16 and PGT145). Conversely, non-NAbs to the CD4-binding site, CD4-induced epitopes or gp41ECTO did not react with the trimers, even when their epitopes were present on simpler forms of Env (e.g. gp120 monomers or dissociated gp41 subunits). Three non-neutralizing MAbs to V3 epitopes did, however, react strongly with the trimers but only by ELISA, and not at all by SPR and to only a limited extent by EM. These new soluble trimers are useful for structural studies and are being assessed for their performance as immunogens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2427.map.gz emd_2427.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2427-v30.xml emd-2427-v30.xml emd-2427.xml emd-2427.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2427.tif EMD-2427.tif | 55.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2427 http://ftp.pdbj.org/pub/emdb/structures/EMD-2427 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2427 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2427 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2427.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2427.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bg505 SOSIP.664 with PGV04 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HIV spike protein 664G construct in complex with Fab fragment of ...
| Entire | Name: HIV spike protein 664G construct in complex with Fab fragment of PGV04 monoclonal antibody |
|---|---|
| Components |
|
-Supramolecule #1000: HIV spike protein 664G construct in complex with Fab fragment of ...
| Supramolecule | Name: HIV spike protein 664G construct in complex with Fab fragment of PGV04 monoclonal antibody type: sample / ID: 1000 Details: The PGV04 Fab was incubated for 40 minutes at 4C with BG505 SOSIP.664. This complex was purified using a size exclusion chromatography and concentrated. Oligomeric state: One trimer binds to three Fabs / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 510 KDa / Theoretical: 510 KDa / Method: Size exclusion Chromatography |
-Macromolecule #1: HIV spike protein BG505 SOSIP.664
| Macromolecule | Name: HIV spike protein BG505 SOSIP.664 / type: protein_or_peptide / ID: 1 / Name.synonym: BG505 SOSIP.664 / Details: BG505 SOSIP.664 was complexed with PGV04 / Number of copies: 1 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus Human immunodeficiency virus |
| Molecular weight | Experimental: 360 KDa / Theoretical: 360 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK293T / Recombinant plasmid: pPPI4 Homo sapiens (human) / Recombinant cell: HEK293T / Recombinant plasmid: pPPI4 |
| Sequence | UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: PGV04 FAB
| Macromolecule | Name: PGV04 FAB / type: protein_or_peptide / ID: 2 Details: produced as an IgG in mammalian cells and digested with papain into Fab Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK293T Homo sapiens (human) / Recombinant cell: HEK293T |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50mM TRIS-HCl 150mM NaCl |
| Staining | Type: NEGATIVE / Details: 2% w/v Uranyl Formate for 25 seconds |
| Grid | Details: 400 mesh copper grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52050 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 1.3 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 52000 Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 1.3 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 50 |
| Temperature | Min: 292 K / Max: 294 K / Average: 293 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification Legacy - Electron beam tilt params: -2 |
| Date | Apr 16, 2013 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 834 / Average electron dose: 55 e/Å2 / Od range: 1.4 |
| Tilt angle min | 0 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final two d classification | Number classes: 256 |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN2, EMAN1 ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN2, EMAN1Details: The particles were selected using a DoG Picker, and cleaned using reference free class averaging. The final map was calculated from a single dataset Number images used: 32867 |
| Details | Particles were picked automatically using DoG Picker and put into a particle stack using the Appion software package Initial, reference-free, two-dimensional (2D) class averages were calculated using particles binned by five via the Xmipp Clustering 2D Alignment and sorted into classes. EMAN was used for the 3D reconstruction. |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation, Fit in Map, Chimera |
 Movie
Movie Controller
Controller