[English] 日本語
 Yorodumi
Yorodumi- EMDB-2158: Bacterial chemoreceptor arrays are hexagonally packed trimers of ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2158 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
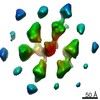
| Title | Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins | |||||||||
 Map data Map data | 3-fold average of wild-type E.coli (strain RP437) Chemoreceptor array sub-volumes (composed of 6 trimers-of receptor dimers) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  chemotaxis / chemotaxis /  signal transduction / signal transduction /  two-component systems two-component systems | |||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
| Method | subtomogram averaging /  cryo EM cryo EM | |||||||||
 Authors Authors | Briegel A / Li X / Bilwes AM / Hughes KT / Jensen GJ / Crane BR | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2012 Journal: Proc Natl Acad Sci U S A / Year: 2012Title: Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Authors: Ariane Briegel / Xiaoxiao Li / Alexandrine M Bilwes / Kelly T Hughes / Grant J Jensen / Brian R Crane /  Abstract: Chemoreceptor arrays are supramolecular transmembrane machines of unknown structure that allow bacteria to sense their surroundings and respond by chemotaxis. We have combined X-ray crystallography ...Chemoreceptor arrays are supramolecular transmembrane machines of unknown structure that allow bacteria to sense their surroundings and respond by chemotaxis. We have combined X-ray crystallography of purified proteins with electron cryotomography of native arrays inside cells to reveal the arrangement of the component transmembrane receptors, histidine kinases (CheA) and CheW coupling proteins. Trimers of receptor dimers lie at the vertices of a hexagonal lattice in a "two-facing-two" configuration surrounding a ring of alternating CheA regulatory domains (P5) and CheW couplers. Whereas the CheA kinase domains (P4) project downward below the ring, the CheA dimerization domains (P3) link neighboring rings to form an extended, stable array. This highly interconnected protein architecture underlies the remarkable sensitivity and cooperative nature of transmembrane signaling in bacterial chemotaxis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2158.map.gz emd_2158.map.gz | 223.1 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2158-v30.xml emd-2158-v30.xml emd-2158.xml emd-2158.xml | 11 KB 11 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2158.png EMD-2158.png | 74.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2158 http://ftp.pdbj.org/pub/emdb/structures/EMD-2158 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2158 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2158 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2158.map.gz / Format: CCP4 / Size: 297.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2158.map.gz / Format: CCP4 / Size: 297.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3-fold average of wild-type E.coli (strain RP437) Chemoreceptor array sub-volumes (composed of 6 trimers-of receptor dimers) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.48 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacterial chemoreceptor arrays
| Entire | Name: Bacterial chemoreceptor arrays |
|---|---|
| Components |
|
-Supramolecule #1000: Bacterial chemoreceptor arrays
| Supramolecule | Name: Bacterial chemoreceptor arrays / type: sample / ID: 1000 Oligomeric state: Trimers of receptor dimers lie at the vertices of a hexagonal lattice in a two-facing-two configuration surrounding a ring of alternating CheA regulatory domains (P5) and CheW couplers Number unique components: 3 |
|---|
-Macromolecule #1: cheA
| Macromolecule | Name: cheA / type: protein_or_peptide / ID: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) / Strain: MG1655 / Location in cell: Inner membrane Escherichia coli (E. coli) / Strain: MG1655 / Location in cell: Inner membrane |
-Macromolecule #2: cheW
| Macromolecule | Name: cheW / type: protein_or_peptide / ID: 2 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) / Strain: MG1655 / Location in cell: Inner membrane Escherichia coli (E. coli) / Strain: MG1655 / Location in cell: Inner membrane |
-Macromolecule #3: Chemoreceptors
| Macromolecule | Name: Chemoreceptors / type: protein_or_peptide / ID: 3 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) / Strain: MG1655 / Location in cell: Inner membrane Escherichia coli (E. coli) / Strain: MG1655 / Location in cell: Inner membrane |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
- Sample preparation
Sample preparation
| Buffer | Details: Tryptone Broth (10 g Tryptone, 5 g NaCl per liter). Cells were grown to exponential phase and incubated with penicillin for 1 hour prior to freezing |
|---|---|
| Grid | Details: R 2/2 copper/Rhodium grids, glow discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.2 mm / Nominal defocus min: 10.0 µm / Nominal magnification: 34000 Bright-field microscopy / Cs: 2.2 mm / Nominal defocus min: 10.0 µm / Nominal magnification: 34000 |
| Specialist optics | Energy filter - Name: Gif 2002 hybrid (Gatan) / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Sample stage | Specimen holder model: OTHER / Tilt series - Axis1 - Min angle: -70 ° / Tilt series - Axis1 - Max angle: 58 ° |
| Temperature | Average: 77 K |
| Date | Jan 15, 2008 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 150 e/Å2 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: imod |
|---|---|
| Final reconstruction | Software - Name:  imod imod |
| Details | CTF correction in imod. Average number of tilts used in the 3D reconstructions: 130. Average tomographic tilt angle increment: 1. |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D |
|---|---|
| Details | Protocol: rigid body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller