[English] 日本語
 Yorodumi
Yorodumi- EMDB-21121: cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21121 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2) | |||||||||
 Map data Map data | half map 2 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationradial glia guided migration of Purkinje cell / cerebral cortex radially oriented cell migration / ERBB2 signaling pathway /  Neddylation / Antigen processing: Ubiquitination & Proteasome degradation / cullin-RING ubiquitin ligase complex / protein neddylation / NEDD8 ligase activity / IgG binding / Cul5-RING ubiquitin ligase complex ...radial glia guided migration of Purkinje cell / cerebral cortex radially oriented cell migration / ERBB2 signaling pathway / Neddylation / Antigen processing: Ubiquitination & Proteasome degradation / cullin-RING ubiquitin ligase complex / protein neddylation / NEDD8 ligase activity / IgG binding / Cul5-RING ubiquitin ligase complex ...radial glia guided migration of Purkinje cell / cerebral cortex radially oriented cell migration / ERBB2 signaling pathway /  Neddylation / Antigen processing: Ubiquitination & Proteasome degradation / cullin-RING ubiquitin ligase complex / protein neddylation / NEDD8 ligase activity / IgG binding / Cul5-RING ubiquitin ligase complex / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / Neddylation / Antigen processing: Ubiquitination & Proteasome degradation / cullin-RING ubiquitin ligase complex / protein neddylation / NEDD8 ligase activity / IgG binding / Cul5-RING ubiquitin ligase complex / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process /  SCF ubiquitin ligase complex / intrinsic apoptotic signaling pathway in response to oxidative stress / apoptotic mitochondrial changes / cullin family protein binding / site of DNA damage / intrinsic apoptotic signaling pathway / Vif-mediated degradation of APOBEC3G / G1/S transition of mitotic cell cycle / SCF ubiquitin ligase complex / intrinsic apoptotic signaling pathway in response to oxidative stress / apoptotic mitochondrial changes / cullin family protein binding / site of DNA damage / intrinsic apoptotic signaling pathway / Vif-mediated degradation of APOBEC3G / G1/S transition of mitotic cell cycle /  calcium channel activity / Inactivation of CSF3 (G-CSF) signaling / Downregulation of ERBB2 signaling / ubiquitin-protein transferase activity / activation of cysteine-type endopeptidase activity involved in apoptotic process / calcium channel activity / Inactivation of CSF3 (G-CSF) signaling / Downregulation of ERBB2 signaling / ubiquitin-protein transferase activity / activation of cysteine-type endopeptidase activity involved in apoptotic process /  ubiquitin protein ligase activity / protein-macromolecule adaptor activity / Antigen processing: Ubiquitination & Proteasome degradation / ubiquitin protein ligase activity / protein-macromolecule adaptor activity / Antigen processing: Ubiquitination & Proteasome degradation /  signaling receptor activity / signaling receptor activity /  Neddylation / ubiquitin-dependent protein catabolic process / protein ubiquitination / Neddylation / ubiquitin-dependent protein catabolic process / protein ubiquitination /  ubiquitin protein ligase binding / negative regulation of apoptotic process / zinc ion binding / extracellular region / ubiquitin protein ligase binding / negative regulation of apoptotic process / zinc ion binding / extracellular region /  nucleoplasm / nucleoplasm /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 5.2 Å cryo EM / Resolution: 5.2 Å | |||||||||
 Authors Authors | Komives EA / Lumpkin RJ / Baker RW / Leschziner AE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structure and dynamics of the ASB9 CUL-RING E3 Ligase. Authors: Ryan J Lumpkin / Richard W Baker / Andres E Leschziner / Elizabeth A Komives /  Abstract: The Cullin 5 (CUL5) Ring E3 ligase uses adaptors Elongins B and C (ELOB/C) to bind different SOCS-box-containing substrate receptors, determining the substrate specificity of the ligase. The 18- ...The Cullin 5 (CUL5) Ring E3 ligase uses adaptors Elongins B and C (ELOB/C) to bind different SOCS-box-containing substrate receptors, determining the substrate specificity of the ligase. The 18-member ankyrin and SOCS box (ASB) family is the largest substrate receptor family. Here we report cryo-EM data for the substrate, creatine kinase (CKB) bound to ASB9-ELOB/C, and for full-length CUL5 bound to the RING protein, RBX2, which binds various E2s. To date, no full structures are available either for a substrate-bound ASB nor for CUL5. Hydrogen-deuterium exchange (HDX-MS) mapped onto a full structural model of the ligase revealed long-range allostery extending from the substrate through CUL5. We propose a revised allosteric mechanism for how CUL-E3 ligases function. ASB9 and CUL5 behave as rigid rods, connected through a hinge provided by ELOB/C transmitting long-range allosteric crosstalk from the substrate through CUL5 to the RBX2 flexible linker. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21121.map.gz emd_21121.map.gz | 28.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21121-v30.xml emd-21121-v30.xml emd-21121.xml emd-21121.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21121.png emd_21121.png | 28.6 KB | ||
| Others |  emd_21121_additional.map.gz emd_21121_additional.map.gz emd_21121_half_map_1.map.gz emd_21121_half_map_1.map.gz emd_21121_half_map_2.map.gz emd_21121_half_map_2.map.gz | 15.2 MB 28.4 MB 28.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21121 http://ftp.pdbj.org/pub/emdb/structures/EMD-21121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21121 | HTTPS FTP |
-Related structure data
| Related structure data |  6v9iMC  6v9hC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21121.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21121.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unfiltered map
| File | emd_21121_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
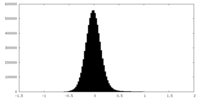
| Density Histograms |
-Half map: half map 1
| File | emd_21121_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
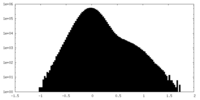
| Density Histograms |
-Half map: unfiltered map
| File | emd_21121_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2)
| Entire | Name: cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2) |
|---|---|
| Components |
|
-Supramolecule #1: cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2)
| Supramolecule | Name: cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) / Recombinant strain: BL21 Escherichia coli (E. coli) / Recombinant strain: BL21 |
| Molecular weight | Theoretical: 100 KDa |
-Macromolecule #1: Immunoglobulin G-binding protein G,Cullin-5
| Macromolecule | Name: Immunoglobulin G-binding protein G,Cullin-5 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 100.4565 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21 (bacteria) Escherichia coli BL21 (bacteria) |
| Sequence | String: MGSSHHHHHH SQDPMEYKLI LNGKTLKGET TTEAVDAATA EKVFKQYAND NGVDGEWTYD DATKTFTVTE IPTTENLYFQ GEFMATSNL LKNKGSLQFE DKWDFMRPIV LKLLRQESVT KQQWFDLFSD VHAVCLWDDK GPAKIHQALK EDILEFIKQA Q ARVLSHQD ...String: MGSSHHHHHH SQDPMEYKLI LNGKTLKGET TTEAVDAATA EKVFKQYAND NGVDGEWTYD DATKTFTVTE IPTTENLYFQ GEFMATSNL LKNKGSLQFE DKWDFMRPIV LKLLRQESVT KQQWFDLFSD VHAVCLWDDK GPAKIHQALK EDILEFIKQA Q ARVLSHQD DTALLKAYIV EWRKFFTQCD ILPKPFCQLE ITLMGKQGSN KKSNVEDSIV RKLMLDTWNE SIFSNIKNRL QD SAMKLVH AERLGEAFDS QLVIGVRESY VNLCSNPEDK LQIYRDNFEK AYLDSTERFY RTQAPSYLQQ NGVQNYMKYA DAK LKEEEK RALRYLETRR ECNSVEALME CCVNALVTSF KETILAECQG MIKRNETEKL HLMFSLMDKV PNGIEPMLKD LEEH IISAG LADMVAAAET ITTDSEKYVE QLLTLFNRFS KLVKEAFQDD PRFLTARDKA YKAVVNDATI FKLELPLKQK GVGLK TQPE SKCPELLANY CDMLLRKTPL SKKLTSEEIE AKLKEVLLVL KYVQNKDVFM RYHKAHLTRR LILDISADSE IEENMV EWL REVGMPADYV NKLARMFQDI KVSEDLNQAF KEMHKNNKLA LPADSVNIKI LNAGAWSRSS EKVFVSLPTE LEDLIPE VE EFYKKNHSGR KLHWHHLMSN GIITFKNEVG QYDLEVTTFQ LAVLFAWNQR PREKISFENL KLATELPDAE LRRTLWSL V AFPKLKRQVL LYEPQVNSPK DFTEGTLFSV NQEFSLIKNA KVQKRGKINL IGRLQLTTER MREEENEGIV QLRILRTQE AIIQIMKMRK KISNAQLQTE LVEILKNMFL PQKKMIKEQI EWLIEHKYIR RDESDINTFI YMA |
-Macromolecule #2: RING-box protein 2
| Macromolecule | Name: RING-box protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 12.7215 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MADVEDGEEP CVLSSHSGSA GSKSGGDKMF SLKKWNAVAM WSWDVECDTC AICRVQVMDA CLRCQAENKQ EDCVVVWGEC NHSFHNCCM SLWVKQNNRC PLCQQDWVVQ RIGK |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 4 second blot time, blot force 20. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 36000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 36000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 8.0 sec. / Average electron dose: 59.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: CTFFIND (ver. 4) |
|---|---|
| Startup model | Type of model: OTHER / Details: Ab initio model generation in cryoSPARC v2 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER / Details: Non-uniform refinement in cryoSPARC v2. |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 5.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2) / Details: Non-uniform refinement in cryoSPARC v2 / Number images used: 46877 |
 Movie
Movie Controller
Controller
















 Z
Z Y
Y X
X