[English] 日本語
 Yorodumi
Yorodumi- EMDB-1970: Negative stained image reconstruction of HIV spike protein in com... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1970 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
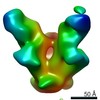
| Title | Negative stained image reconstruction of HIV spike protein in complex with PGT128 Fab at 14 Angstrom resolution | |||||||||
 Map data Map data | HIV Envelope protein in complex with Fab PGT128 Env (gene) Env (gene) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  HIV / HIV /  broadly neutralizing antibody / glycan shield broadly neutralizing antibody / glycan shield | |||||||||
| Biological species |    Human immunodeficiency virus / Human immunodeficiency virus /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 14.0 Å negative staining / Resolution: 14.0 Å | |||||||||
 Authors Authors | Khayat R / Pejchal R / Julien J-P / Wilson IA | |||||||||
 Citation Citation |  Journal: Science / Year: 2011 Journal: Science / Year: 2011Title: A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Authors: Robert Pejchal / Katie J Doores / Laura M Walker / Reza Khayat / Po-Ssu Huang / Sheng-Kai Wang / Robyn L Stanfield / Jean-Philippe Julien / Alejandra Ramos / Max Crispin / Rafael Depetris / ...Authors: Robert Pejchal / Katie J Doores / Laura M Walker / Reza Khayat / Po-Ssu Huang / Sheng-Kai Wang / Robyn L Stanfield / Jean-Philippe Julien / Alejandra Ramos / Max Crispin / Rafael Depetris / Umesh Katpally / Andre Marozsan / Albert Cupo / Sebastien Maloveste / Yan Liu / Ryan McBride / Yukishige Ito / Rogier W Sanders / Cassandra Ogohara / James C Paulson / Ten Feizi / Christopher N Scanlan / Chi-Huey Wong / John P Moore / William C Olson / Andrew B Ward / Pascal Poignard / William R Schief / Dennis R Burton / Ian A Wilson /  Abstract: The HIV envelope (Env) protein gp120 is protected from antibody recognition by a dense glycan shield. However, several of the recently identified PGT broadly neutralizing antibodies appear to ...The HIV envelope (Env) protein gp120 is protected from antibody recognition by a dense glycan shield. However, several of the recently identified PGT broadly neutralizing antibodies appear to interact directly with the HIV glycan coat. Crystal structures of antigen-binding fragments (Fabs) PGT 127 and 128 with Man(9) at 1.65 and 1.29 angstrom resolution, respectively, and glycan binding data delineate a specific high mannose-binding site. Fab PGT 128 complexed with a fully glycosylated gp120 outer domain at 3.25 angstroms reveals that the antibody penetrates the glycan shield and recognizes two conserved glycans as well as a short β-strand segment of the gp120 V3 loop, accounting for its high binding affinity and broad specificity. Furthermore, our data suggest that the high neutralization potency of PGT 127 and 128 immunoglobulin Gs may be mediated by cross-linking Env trimers on the viral surface. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1970.map.gz emd_1970.map.gz | 14.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1970-v30.xml emd-1970-v30.xml emd-1970.xml emd-1970.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| Images |  1970.png 1970.png | 246.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1970 http://ftp.pdbj.org/pub/emdb/structures/EMD-1970 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1970 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1970 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1970.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1970.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HIV Envelope protein in complex with Fab PGT128 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.18 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HIV spike protein 664G construct in complex with Fab fragment of ...
| Entire | Name: HIV spike protein 664G construct in complex with Fab fragment of PGT128 monoclonal antibody |
|---|---|
| Components |
|
-Supramolecule #1000: HIV spike protein 664G construct in complex with Fab fragment of ...
| Supramolecule | Name: HIV spike protein 664G construct in complex with Fab fragment of PGT128 monoclonal antibody type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: One trimer binds to three Fabs / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 445 KDa / Method: Size exclusion chromatography and light scattering |
-Macromolecule #1: Human immunodeficiency virus envelope protein
| Macromolecule | Name: Human immunodeficiency virus envelope protein / type: protein_or_peptide / ID: 1 / Name.synonym: Spike protein Details: The PGT128 Fab was complexed to the spike protein, partially deglycosylated, purified using size exclusion chromatography, and concentrated. Number of copies: 3 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus / Strain: KNH1144 SOSIP 664G / synonym: Spike protien Human immunodeficiency virus / Strain: KNH1144 SOSIP 664G / synonym: Spike protien |
| Recombinant expression | Organism: 293S / Recombinant plasmid: pP14 |
-Macromolecule #2: PGT128 monoclonal antibody fragment
| Macromolecule | Name: PGT128 monoclonal antibody fragment / type: protein_or_peptide / ID: 2 / Name.synonym: Fab fragment / Oligomeric state: Monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM HEPES pH 7.5, 50 mM NaCl |
| Staining | Type: NEGATIVE / Details: 2% Uranyl Formate for 30 seconds |
| Grid | Details: 400 mesh copper grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 0.95 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 100000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 0.95 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 100000 |
| Sample stage | Specimen holder: eucentric / Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 55 |
| Temperature | Min: 293 K / Max: 293 K / Average: 293 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification Legacy - Electron beam tilt params: -2 mrad |
| Date | Aug 11, 2011 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (4k x 4k) / Digitization - Sampling interval: 0.109 µm / Number real images: 357 / Average electron dose: 16 e/Å2 / Details: Data collected on CCD / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Micrograph and each particle |
|---|---|
| Final two d classification | Number classes: 62 |
| Final angle assignment | Details: SPIDER:theta 90 degrees, phi 120 degrees |
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 14.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Sparx / Number images used: 12824 ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 14.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Sparx / Number images used: 12824 |
| Details | The particles were selected using an DoG Picker, and cleaned using reference free class averaging. |
 Movie
Movie Controller
Controller