+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1687 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EP helicase hexamer from Double hexamer of LTag108-627 mutant | |||||||||
 Map data Map data | EP Helicase hexamer from double hexamer LTag108-627 mutant | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Helicase / Helicase /  Large T antigen / Large T antigen /  DNA Replication / DNA Replication /  SV40 SV40 | |||||||||
| Biological species |   Simian virus 40 Simian virus 40 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 19.0 Å cryo EM / Resolution: 19.0 Å | |||||||||
 Authors Authors | Cuesta I / Nunez-Ramirez R / Scheres SHW / Gai D / Chen XS / Fanning E / Carazo JM | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2010 Journal: J Mol Biol / Year: 2010Title: Conformational rearrangements of SV40 large T antigen during early replication events. Authors: Isabel Cuesta / Rafael Núñez-Ramírez / Sjors H W Scheres / Dahai Gai / Xiaojiang S Chen / Ellen Fanning / Jose María Carazo /  Abstract: The Simian virus 40 (SV40) large tumor antigen (LTag) functions as the replicative helicase and initiator for viral DNA replication. For SV40 replication, the first essential step is the assembly of ...The Simian virus 40 (SV40) large tumor antigen (LTag) functions as the replicative helicase and initiator for viral DNA replication. For SV40 replication, the first essential step is the assembly of an LTag double hexamer at the origin DNA that will subsequently melt the origin DNA to initiate fork unwinding. In this study, we used three-dimensional cryo-electron microscopy to visualize early events in the activation of DNA replication in the SV40 model system. We obtained structures of wild-type double-hexamer complexes of LTag bound to SV40 origin DNA, to which atomic structures have been fitted. Wild-type LTag was observed in two distinct conformations: In one conformation, the central module containing the J-domains and the origin binding domains of both hexamers is a compact closed ring. In the other conformation, the central module is an open ring with a gap formed by rearrangement of the N-terminal regions of the two hexamers, potentially allowing for the passage of single-stranded DNA generated from the melted origin DNA. Double-hexamer complexes containing mutant LTag that lacks the N-terminal J-domain show the central module predominantly in the closed-ring state. Analyses of the LTag C-terminal regions reveal that the LTag hexamers bound to the A/T-rich tract origin of replication and early palindrome origin of replication elements are structurally distinct. Lastly, visualization of DNA density protruding from the LTag C-terminal domains suggests that oligomerization of the LTag complex takes place on double-stranded DNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1687.map.gz emd_1687.map.gz | 186.3 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1687-v30.xml emd-1687-v30.xml emd-1687.xml emd-1687.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_1687.jpg emd_1687.jpg | 10 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1687 http://ftp.pdbj.org/pub/emdb/structures/EMD-1687 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1687 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1687 | HTTPS FTP |
-Related structure data
| Related structure data |  1681C  1682C  1683C 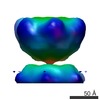 1684C  1685C  1686C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1687.map.gz / Format: CCP4 / Size: 245.1 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1687.map.gz / Format: CCP4 / Size: 245.1 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EP Helicase hexamer from double hexamer LTag108-627 mutant | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : EP helicase hexamer from double hexamer of LTag108-627 mutant
| Entire | Name: EP helicase hexamer from double hexamer of LTag108-627 mutant |
|---|---|
| Components |
|
-Supramolecule #1000: EP helicase hexamer from double hexamer of LTag108-627 mutant
| Supramolecule | Name: EP helicase hexamer from double hexamer of LTag108-627 mutant type: sample / ID: 1000 / Oligomeric state: Hexameric / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 400 KDa / Theoretical: 400 KDa |
-Macromolecule #1: LTag108-627
| Macromolecule | Name: LTag108-627 / type: protein_or_peptide / ID: 1 / Name.synonym: LTag108-627 / Number of copies: 12 / Oligomeric state: Dodecameric / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Simian virus 40 / synonym: SV40 Simian virus 40 / synonym: SV40 |
| Molecular weight | Experimental: 400 KDa / Theoretical: 400 KDa |
-Macromolecule #2: SV40 ori
| Macromolecule | Name: SV40 ori / type: dna / ID: 2 / Name.synonym: SV40 ori / Classification: DNA / Structure: DOUBLE HELIX / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism:   Simian virus 40 / synonym: SV40 Simian virus 40 / synonym: SV40 |
| Molecular weight | Experimental: 70 KDa / Theoretical: 70 KDa |
| Sequence | String: aagctttctc actacttctg gaatagctca gaggccgagg cggcctcggc ctctgcataa ataaaaaaaa ttagtcagcc atgggtcgac cggatccccg |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM TrisHCl (pH 7.5), 100 mM NaCl, 5 mM KCl, 5 mM MgCl2, 3 mM ADP |
| Grid | Details: Quantifoil grids |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 93 K / Instrument: LEICA PLUNGER / Details: Vitrification instrument: Leica Plunger / Method: Blot for 2-3 second before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.26 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 50000 Bright-field microscopy / Cs: 2.26 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Min: 93 K / Max: 93 K / Average: 93 K |
| Alignment procedure | Legacy - Astigmatism: CTF correction |
| Date | Nov 26, 2007 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Number real images: 60 / Average electron dose: 10 e/Å2 / Bits/pixel: 8 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Plate |
|---|---|
| Final two d classification | Number classes: 26 |
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 19.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Xmipp / Number images used: 9000 ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 19.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Xmipp / Number images used: 9000 |
| Details | symmetry C6 was applied in 3D reconstruction. Tilt angle restricted to 60-90 degrees |
 Movie
Movie Controller
Controller










