+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human GBP1 dimer bound to GDP-AlF3 | |||||||||
 Map data Map data | Primary map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cell-autonomous immunity / intracellular pathogens /  GTPase / GTPase /  IMMUNE SYSTEM IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationGDP phosphatase activity / non-canonical inflammasome complex assembly / protein localization to vacuole / negative regulation of substrate adhesion-dependent cell spreading / symbiont cell surface /  cytolysis in another organism / positive regulation of pyroptotic inflammatory response / vesicle membrane / negative regulation of protein localization to plasma membrane / negative regulation of interleukin-2 production ...GDP phosphatase activity / non-canonical inflammasome complex assembly / protein localization to vacuole / negative regulation of substrate adhesion-dependent cell spreading / symbiont cell surface / cytolysis in another organism / positive regulation of pyroptotic inflammatory response / vesicle membrane / negative regulation of protein localization to plasma membrane / negative regulation of interleukin-2 production ...GDP phosphatase activity / non-canonical inflammasome complex assembly / protein localization to vacuole / negative regulation of substrate adhesion-dependent cell spreading / symbiont cell surface /  cytolysis in another organism / positive regulation of pyroptotic inflammatory response / vesicle membrane / negative regulation of protein localization to plasma membrane / negative regulation of interleukin-2 production / negative regulation of T cell receptor signaling pathway / cytolysis in another organism / positive regulation of pyroptotic inflammatory response / vesicle membrane / negative regulation of protein localization to plasma membrane / negative regulation of interleukin-2 production / negative regulation of T cell receptor signaling pathway /  spectrin binding / spectrin binding /  cytokine binding / defense response to protozoan / cytokine binding / defense response to protozoan /  Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / cellular response to interleukin-1 / regulation of protein localization to plasma membrane / regulation of calcium-mediated signaling / G protein activity / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / cellular response to interleukin-1 / regulation of protein localization to plasma membrane / regulation of calcium-mediated signaling / G protein activity /  lipopolysaccharide binding / lipopolysaccharide binding /  Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement /  Hsp90 protein binding / cytoplasmic vesicle membrane / negative regulation of ERK1 and ERK2 cascade / cellular response to type II interferon / GDP binding / Interferon gamma signaling / Hsp90 protein binding / cytoplasmic vesicle membrane / negative regulation of ERK1 and ERK2 cascade / cellular response to type II interferon / GDP binding / Interferon gamma signaling /  actin cytoskeleton / cellular response to tumor necrosis factor / actin cytoskeleton / cellular response to tumor necrosis factor /  actin binding / cytoplasmic vesicle / defense response to virus / defense response to bacterium / actin binding / cytoplasmic vesicle / defense response to virus / defense response to bacterium /  Golgi membrane / Golgi membrane /  innate immune response / innate immune response /  GTPase activity / GTP binding / GTPase activity / GTP binding /  Golgi apparatus / Golgi apparatus /  enzyme binding / protein homodimerization activity / extracellular region / identical protein binding / enzyme binding / protein homodimerization activity / extracellular region / identical protein binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
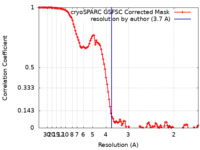
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Kuhm TI / Jakobi AJ | |||||||||
| Funding support | European Union,  Netherlands, 2 items Netherlands, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of the human GBP1 dimer bound to GDP-AlF3 Authors: Kuhm TI / de Agrela Pinto C / Taisne C / Huber S / Giannopolou N / Pardon E / Steyaert J / Jakobi AJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16794.map.gz emd_16794.map.gz | 127.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16794-v30.xml emd-16794-v30.xml emd-16794.xml emd-16794.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16794_fsc.xml emd_16794_fsc.xml | 10.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16794.png emd_16794.png | 100.9 KB | ||
| Masks |  emd_16794_msk_1.map emd_16794_msk_1.map | 134.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16794.cif.gz emd-16794.cif.gz | 7.1 KB | ||
| Others |  emd_16794_additional_1.map.gz emd_16794_additional_1.map.gz emd_16794_half_map_1.map.gz emd_16794_half_map_1.map.gz emd_16794_half_map_2.map.gz emd_16794_half_map_2.map.gz | 1.6 MB 124.9 MB 124.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16794 http://ftp.pdbj.org/pub/emdb/structures/EMD-16794 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16794 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16794 | HTTPS FTP |
-Related structure data
| Related structure data |  8cqbMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16794.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16794.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.834 Å | ||||||||||||||||||||||||||||||||||||
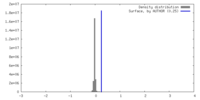
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16794_msk_1.map emd_16794_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
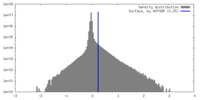
| Density Histograms |
-Additional map: Locally sharpened map (LocScale)
| File | emd_16794_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally sharpened map (LocScale) | ||||||||||||
| Projections & Slices |
| ||||||||||||
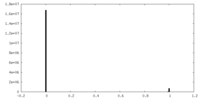
| Density Histograms |
-Half map: Half map 2
| File | emd_16794_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
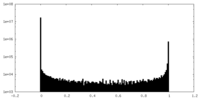
| Density Histograms |
-Half map: Half map 1
| File | emd_16794_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homodimeric complex of GBP1 stabilised by GDP-AlF3
| Entire | Name: Homodimeric complex of GBP1 stabilised by GDP-AlF3 |
|---|---|
| Components |
|
-Supramolecule #1: Homodimeric complex of GBP1 stabilised by GDP-AlF3
| Supramolecule | Name: Homodimeric complex of GBP1 stabilised by GDP-AlF3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 134 KDa |
-Macromolecule #1: Guanylate-binding protein 1
| Macromolecule | Name: Guanylate-binding protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number:  Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 68.021617 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MASEIHMTGP MCLIENTNGR LMANPEALKI LSAITQPMVV VAIVGLYRTG KSYLMNKLAG KKKGFSLGST VQSHTKGIWM WCVPHPKKP GHILVLLDTE GLGDVEKGDN QNDSWIFALA VLLSSTFVYN SIGTINQQAM DQLYYVTELT HRIRSKSSPD E NENEVEDS ...String: MASEIHMTGP MCLIENTNGR LMANPEALKI LSAITQPMVV VAIVGLYRTG KSYLMNKLAG KKKGFSLGST VQSHTKGIWM WCVPHPKKP GHILVLLDTE GLGDVEKGDN QNDSWIFALA VLLSSTFVYN SIGTINQQAM DQLYYVTELT HRIRSKSSPD E NENEVEDS ADFVSFFPDF VWTLRDFSLD LEADGQPLTP DEYLTYSLKL KKGTSQKDET FNLPRLCIRK FFPKKKCFVF DR PVHRRKL AQLEKLQDEE LDPEFVQQVA DFCSYIFSNS KTKTLSGGIQ VNGPRLESLV LTYVNAISSG DLPCMENAVL ALA QIENSA AVQKAIAHYE QQMGQKVQLP TETLQELLDL HRDSEREAIE VFIRSSFKDV DHLFQKELAA QLEKKRDDFC KQNQ EASSD RCSALLQVIF SPLEEEVKAG IYSKPGGYRL FVQKLQDLKK KYYEEPRKGI QAEEILQTYL KSKESMTDAI LQTDQ TLTE KEKEIEVERV KAESAQASAK MLQEMQRKNE QMMEQKERSY QEHLKQLTEK MENDRVQLLK EQERTLALKL QEQEQL LKE GFQKESRIMK NEIQDLQTKM RRRKACTIS UniProtKB: Guanylate-binding protein 1 |
-Macromolecule #2: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #3: ALUMINUM FLUORIDE
| Macromolecule | Name: ALUMINUM FLUORIDE / type: ligand / ID: 3 / Number of copies: 2 / Formula: AF3 |
|---|---|
| Molecular weight | Theoretical: 83.977 Da |
| Chemical component information |  ChemComp-AF3: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 5u mM HEPES pH7.4, 150 mM NaCl, 1 mM DTT, 200 uM GDP, 10 mM NaF, 300 uM AlCl3, 5 mM MgCl2 | ||||||||||||||||||||||||
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % / Chamber temperature: 22 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 5214 / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Target criteria: Real-space cross correlation | ||||||||
| Output model |  PDB-8cqb: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)