[English] 日本語
 Yorodumi
Yorodumi- EMDB-16000: Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
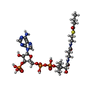
| Title | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane core) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationplastid outer membrane / TIM22 mitochondrial import inner membrane insertion complex / anther dehiscence / vegetative to reproductive phase transition of meristem /  chloroplast outer membrane / chloroplast outer membrane /  NADH dehydrogenase complex / P450-containing electron transport chain / NADH dehydrogenase complex / P450-containing electron transport chain /  Lyases; Carbon-oxygen lyases; Hydro-lyases / protein insertion into mitochondrial inner membrane / Lyases; Carbon-oxygen lyases; Hydro-lyases / protein insertion into mitochondrial inner membrane /  photorespiration ...plastid outer membrane / TIM22 mitochondrial import inner membrane insertion complex / anther dehiscence / vegetative to reproductive phase transition of meristem / photorespiration ...plastid outer membrane / TIM22 mitochondrial import inner membrane insertion complex / anther dehiscence / vegetative to reproductive phase transition of meristem /  chloroplast outer membrane / chloroplast outer membrane /  NADH dehydrogenase complex / P450-containing electron transport chain / NADH dehydrogenase complex / P450-containing electron transport chain /  Lyases; Carbon-oxygen lyases; Hydro-lyases / protein insertion into mitochondrial inner membrane / Lyases; Carbon-oxygen lyases; Hydro-lyases / protein insertion into mitochondrial inner membrane /  photorespiration / embryo development ending in seed dormancy / response to abscisic acid / plant-type vacuole / respiratory chain complex I / photorespiration / embryo development ending in seed dormancy / response to abscisic acid / plant-type vacuole / respiratory chain complex I /  ubiquinone binding / regulation of reactive oxygen species metabolic process / electron transport coupled proton transport / ubiquinone binding / regulation of reactive oxygen species metabolic process / electron transport coupled proton transport /  plastid / plastid /  porin activity / porin activity /  NADH:ubiquinone reductase (H+-translocating) / pore complex / mitochondrial respiratory chain complex I / mitochondrial electron transport, NADH to ubiquinone / protein homotrimerization / NADH:ubiquinone reductase (H+-translocating) / pore complex / mitochondrial respiratory chain complex I / mitochondrial electron transport, NADH to ubiquinone / protein homotrimerization /  NADH dehydrogenase (ubiquinone) activity / ATP synthesis coupled electron transport / NADH dehydrogenase (ubiquinone) activity / ATP synthesis coupled electron transport /  respirasome / monoatomic ion transport / respirasome / monoatomic ion transport /  aerobic respiration / response to salt stress / aerobic respiration / response to salt stress /  chloroplast / chloroplast /  mitochondrial membrane / mitochondrial membrane /  carbonate dehydratase activity / carbonate dehydratase activity /  mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding /  peroxisome / peroxisome /  mitochondrial inner membrane / mitochondrial outer membrane / copper ion binding / mitochondrial inner membrane / mitochondrial outer membrane / copper ion binding /  nucleolus / protein homodimerization activity / nucleolus / protein homodimerization activity /  mitochondrion / extracellular region / identical protein binding / mitochondrion / extracellular region / identical protein binding /  metal ion binding / metal ion binding /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Arabidopsis t (thale cress) / Arabidopsis t (thale cress) /   thale cress (thale cress) thale cress (thale cress) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.13 Å cryo EM / Resolution: 2.13 Å | |||||||||
 Authors Authors | Klusch N / Kuehlbrandt W | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2023 Journal: Nat Plants / Year: 2023Title: Cryo-EM structure of the respiratory I + III supercomplex from Arabidopsis thaliana at 2 Å resolution. Authors: Niklas Klusch / Maximilian Dreimann / Jennifer Senkler / Nils Rugen / Werner Kühlbrandt / Hans-Peter Braun /  Abstract: Protein complexes of the mitochondrial respiratory chain assemble into respiratory supercomplexes. Here we present the high-resolution electron cryo-microscopy structure of the Arabidopsis ...Protein complexes of the mitochondrial respiratory chain assemble into respiratory supercomplexes. Here we present the high-resolution electron cryo-microscopy structure of the Arabidopsis respiratory supercomplex consisting of complex I and a complex III dimer, with a total of 68 protein subunits and numerous bound cofactors. A complex I-ferredoxin, subunit B14.7 and P9, a newly defined subunit of plant complex I, mediate supercomplex formation. The component complexes stabilize one another, enabling new detailed insights into their structure. We describe (1) an interrupted aqueous passage for proton translocation in the membrane arm of complex I; (2) a new coenzyme A within the carbonic anhydrase module of plant complex I defining a second catalytic centre; and (3) the water structure at the proton exit pathway of complex III with a co-purified ubiquinone in the Q site. We propose that the main role of the plant supercomplex is to stabilize its components in the membrane. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16000.map.gz emd_16000.map.gz | 91.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16000-v30.xml emd-16000-v30.xml emd-16000.xml emd-16000.xml | 42.4 KB 42.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16000_fsc.xml emd_16000_fsc.xml | 26.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_16000.png emd_16000.png | 151.6 KB | ||
| Others |  emd_16000_additional_1.map.gz emd_16000_additional_1.map.gz emd_16000_half_map_1.map.gz emd_16000_half_map_1.map.gz emd_16000_half_map_2.map.gz emd_16000_half_map_2.map.gz | 94.9 MB 1.1 GB 1.1 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16000 http://ftp.pdbj.org/pub/emdb/structures/EMD-16000 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16000 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16000 | HTTPS FTP |
-Related structure data
| Related structure data |  8befMC  8bedC  8beeC  8behC  8belC  8bepC  8bpxC  8bq5C  8bq6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16000.map.gz / Format: CCP4 / Size: 98.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16000.map.gz / Format: CCP4 / Size: 98.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.573 Å | ||||||||||||||||||||||||||||||||||||
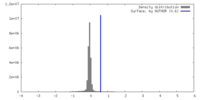
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_16000_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
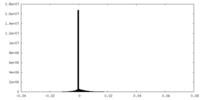
| Density Histograms |
-Half map: #2
| File | emd_16000_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
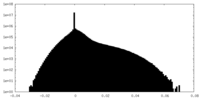
| Density Histograms |
-Half map: #1
| File | emd_16000_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
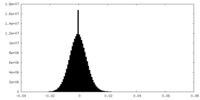
| Density Histograms |
- Sample components
Sample components
+Entire : Mitochondrial Arabidopsis thaliana I+III2 supercomplex (CI membra...
+Supramolecule #1: Mitochondrial Arabidopsis thaliana I+III2 supercomplex (CI membra...
+Macromolecule #1: NADH-ubiquinone oxidoreductase chain 3
+Macromolecule #2: NADH-ubiquinone oxidoreductase chain 1
+Macromolecule #3: NADH-ubiquinone oxidoreductase chain 6
+Macromolecule #4: NADH-ubiquinone oxidoreductase chain 4L
+Macromolecule #5: NADH-ubiquinone oxidoreductase chain 5
+Macromolecule #6: NADH-ubiquinone oxidoreductase chain 4
+Macromolecule #7: NADH-ubiquinone oxidoreductase chain 2
+Macromolecule #8: AT3G07480.1
+Macromolecule #9: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8-B
+Macromolecule #10: Outer envelope pore protein 16-3, chloroplastic/mitochondrial
+Macromolecule #11: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-A
+Macromolecule #12: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1
+Macromolecule #13: At2g46540/F11C10.23
+Macromolecule #14: Excitatory amino acid transporter
+Macromolecule #15: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B
+Macromolecule #16: At4g16450
+Macromolecule #17: P1
+Macromolecule #18: Uncharacterized protein At1g67785
+Macromolecule #19: Uncharacterized protein At2g27730, mitochondrial
+Macromolecule #20: Gamma carbonic anhydrase-like 2, mitochondrial
+Macromolecule #21: Gamma carbonic anhydrase 2, mitochondrial
+Macromolecule #22: Gamma carbonic anhydrase 1, mitochondrial
+Macromolecule #23: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
+Macromolecule #24: Ubiquinone-9
+Macromolecule #25: PHOSPHATIDYLETHANOLAMINE
+Macromolecule #26: 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl...
+Macromolecule #27: FE (III) ION
+Macromolecule #28: 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14...
+Macromolecule #29: (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5...
+Macromolecule #30: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE
+Macromolecule #31: CARDIOLIPIN
+Macromolecule #32: ZINC ION
+Macromolecule #33: Butyryl Coenzyme A
+Macromolecule #34: water
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.18 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: CONTINUOUS / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: GRAPHENE / Support film - #1 - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 215000 Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 215000 |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Software | Name:  Coot (ver. 0.9.5) Coot (ver. 0.9.5) |
|---|---|
| Refinement | Space: REAL |
| Output model |  PDB-8bef: |
 Movie
Movie Controller
Controller



























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)