[English] 日本語
 Yorodumi
Yorodumi- EMDB-15726: Poliovirus type 3 (strain Saukett) stabilised virus-like particle... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and Pleconaril | |||||||||
 Map data Map data | Local resolution scaled final map from RELION. Contour level determined in UCSF ChimeraX. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont genome entry into host cell via pore formation in plasma membrane /  viral capsid / symbiont-mediated suppression of host gene expression / virion attachment to host cell / structural molecule activity viral capsid / symbiont-mediated suppression of host gene expression / virion attachment to host cell / structural molecule activitySimilarity search - Function | |||||||||
| Biological species |   Human poliovirus 3 Human poliovirus 3 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.6 Å cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Bahar MW / Fry EE / Stuart DI | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation. Authors: Mohammad W Bahar / Veronica Nasta / Helen Fox / Lee Sherry / Keith Grehan / Claudine Porta / Andrew J Macadam / Nicola J Stonehouse / David J Rowlands / Elizabeth E Fry / David I Stuart /   Abstract: Strategies to prevent the recurrence of poliovirus (PV) after eradication may utilise non-infectious, recombinant virus-like particle (VLP) vaccines. Despite clear advantages over inactivated or ...Strategies to prevent the recurrence of poliovirus (PV) after eradication may utilise non-infectious, recombinant virus-like particle (VLP) vaccines. Despite clear advantages over inactivated or attenuated virus vaccines, instability of VLPs can compromise their immunogenicity. Glutathione (GSH), an important cellular reducing agent, is a crucial co-factor for the morphogenesis of enteroviruses, including PV. We report cryo-EM structures of GSH bound to PV serotype 3 VLPs showing that it can enhance particle stability. GSH binds the positively charged pocket at the interprotomer interface shown recently to bind GSH in enterovirus F3 and putative antiviral benzene sulphonamide compounds in other enteroviruses. We show, using high-resolution cryo-EM, the binding of a benzene sulphonamide compound with a PV serotype 2 VLP, consistent with antiviral activity through over-stabilizing the interprotomer pocket, preventing the capsid rearrangements necessary for viral infection. Collectively, these results suggest GSH or an analogous tight-binding antiviral offers the potential for stabilizing VLP vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15726.map.gz emd_15726.map.gz | 195.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15726-v30.xml emd-15726-v30.xml emd-15726.xml emd-15726.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15726_fsc.xml emd_15726_fsc.xml | 15.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_15726.png emd_15726.png | 93.4 KB | ||
| Others |  emd_15726_half_map_1.map.gz emd_15726_half_map_1.map.gz emd_15726_half_map_2.map.gz emd_15726_half_map_2.map.gz | 275.9 MB 276 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15726 http://ftp.pdbj.org/pub/emdb/structures/EMD-15726 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15726 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15726 | HTTPS FTP |
-Related structure data
| Related structure data |  8ayyMC  8ayxC  8ayzC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15726.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15726.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution scaled final map from RELION. Contour level determined in UCSF ChimeraX. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map from final refinement in RELION. Contour...
| File | emd_15726_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map from final refinement in RELION. Contour level determined in UCSF ChimeraX. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map from final refinement in RELION. Contour...
| File | emd_15726_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map from final refinement in RELION. Contour level determined in UCSF ChimeraX. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human poliovirus 3
| Entire | Name:   Human poliovirus 3 Human poliovirus 3 |
|---|---|
| Components |
|
-Supramolecule #1: Human poliovirus 3
| Supramolecule | Name: Human poliovirus 3 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Recombinantly expressed virus-like particle of poliovirus type 3 (strain Saukett). NCBI-ID: 12086 / Sci species name: Human poliovirus 3 / Sci species strain: Saukett / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5.85 MDa |
| Virus shell | Shell ID: 1 / Name: Virus shell 1 / Diameter: 310.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein, VP1
| Macromolecule | Name: Capsid protein, VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human poliovirus 3 Human poliovirus 3 |
| Molecular weight | Theoretical: 33.562785 KDa |
| Recombinant expression | Organism:   Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: GIEDLITEVA QGALTLSLPK QQDSLPDTKA SGPAHSKEVP ALTAVETGAT NPLVPSDTVQ TRHVIQRRSR SESTIESFFA RGACVAIIE VDNEEPTTRA QKLFAMWRIT YKDTVQLRRK LEFFTYSRFD MELTFVVTAN FTNTNNGHAL NQVYQIMYIP P GAPTPKSW ...String: GIEDLITEVA QGALTLSLPK QQDSLPDTKA SGPAHSKEVP ALTAVETGAT NPLVPSDTVQ TRHVIQRRSR SESTIESFFA RGACVAIIE VDNEEPTTRA QKLFAMWRIT YKDTVQLRRK LEFFTYSRFD MELTFVVTAN FTNTNNGHAL NQVYQIMYIP P GAPTPKSW DDYTWQTSSN PSIFYTYGAA PARISVPYVG LANAYSHFYD GFAKVPLKTD ANDQIGDSLY SAMTVDDFGV LA IRVVNDH NPTKVTSKVR IYMKPKHVRV WCPRPPRAVP YYGPGVDYKD NLNPLSEKGL TTY |
-Macromolecule #2: Capsid protein, VP0
| Macromolecule | Name: Capsid protein, VP0 / type: protein_or_peptide / ID: 2 Details: Sequence is given for the VP0 polypeptide. Mutations are numbered according to sequence numbering for mature polypeptides VP2 and VP4. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human poliovirus 3 Human poliovirus 3 |
| Molecular weight | Theoretical: 37.623023 KDa |
| Recombinant expression | Organism:   Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MGAQVSSQKV GAHENSNRAY GGSTINYTTI NYYKDSASNA ASKQDYSQDP SKFTEPLKDV LIKTAPALNS PNVEACGYSD RVLQLTIGN STITTQEAAN SVVAYGRWPE FIRDDEANPV DQPTEPDVAT CRFYTLDTVM WGKESKGWWW KLPDALRDMG L FGQNMYYH ...String: MGAQVSSQKV GAHENSNRAY GGSTINYTTI NYYKDSASNA ASKQDYSQDP SKFTEPLKDV LIKTAPALNS PNVEACGYSD RVLQLTIGN STITTQEAAN SVVAYGRWPE FIRDDEANPV DQPTEPDVAT CRFYTLDTVM WGKESKGWWW KLPDALRDMG L FGQNMYYH YLGRSGYTVH VQCNASKFHQ GALGVFAIPE YCLAGDSDKQ RYTSYANANP GEKGGKFYSQ FNRDTAVTSP KR EFCPVDY LLGCGVLLGN AFVYPHQIIN LRTNNSATIV LPYVNAMAID SMVKHNNWGI AILPLSPLDF AQESSVEIPI TVT IAPMCS EFNGLRNVTA PKFQ |
-Macromolecule #3: Capsid protein, VP3
| Macromolecule | Name: Capsid protein, VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human poliovirus 3 Human poliovirus 3 |
| Molecular weight | Theoretical: 26.3151 KDa |
| Recombinant expression | Organism:   Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: GLPVLNTPGS NQYLTSDNYQ SPCAIPEFDV TPPIDIPGEV KNMMELAEID TMIPLNLENT KRNTMDMYRV TLSDSADLSQ PILCFSLSP ASDPRLSHTM LGEVLNYYTH WAGSLKFTFL FCGSMMATGK ILVAYAPPGA QPPTSRKEAM LGTHVIWDLG L QSSCTMVV ...String: GLPVLNTPGS NQYLTSDNYQ SPCAIPEFDV TPPIDIPGEV KNMMELAEID TMIPLNLENT KRNTMDMYRV TLSDSADLSQ PILCFSLSP ASDPRLSHTM LGEVLNYYTH WAGSLKFTFL FCGSMMATGK ILVAYAPPGA QPPTSRKEAM LGTHVIWDLG L QSSCTMVV PWISNVTYRQ TTQDSFTEGG YISMFYQTRI VVPLSTPKSM SMLGFVSACN DFSVRLLRDT THISQSALPQ |
-Macromolecule #4: 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-...
| Macromolecule | Name: 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE type: ligand / ID: 4 / Number of copies: 1 / Formula: W11 |
|---|---|
| Molecular weight | Theoretical: 381.349 Da |
| Chemical component information | 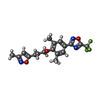 ChemComp-W11: |
-Macromolecule #5: GLUTATHIONE
| Macromolecule | Name: GLUTATHIONE / type: ligand / ID: 5 / Number of copies: 1 / Formula: GSH |
|---|---|
| Molecular weight | Theoretical: 307.323 Da |
| Chemical component information |  ChemComp-GSH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
Details: 1 x DPBS, 20 mM EDTA, pH 7.0 | ||||||
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Support film - Film thickness: 3.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR Details: The specific type of grid used was Ultra-thin carbon support film, 3nm - on lacey carbon AGS187-4 from Agar Scientific. | ||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: 4 ul of sample blotted for 3.5 seconds with -15 blot force on FEI Vitrobot mark IV.. | ||||||
| Details | Virus-like particle purified by sucrose density gradient purification from Pichia pastoris cells. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 129629 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.9 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 75000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.9 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 4706 / Average exposure time: 1.15 sec. / Average electron dose: 35.29 e/Å2 / Details: Pixel sampling of 1.08 A/pixel. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Details | Initial model was rigid body fitted using UCSF chimera and Coot. Global minimization and B-factor refinement was performed in real space using phenix_real.space.refine. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
| Output model |  PDB-8ayy: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)







































