+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AQP7 dimer of tetramers_D4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationTransport of glycerol from adipocytes to the liver by Aquaporins / glycerol channel activity / urea transmembrane transporter activity / Passive transport by Aquaporins / glycerol transmembrane transport /  water channel activity / water transport / water channel activity / water transport /  lipid droplet / cytoplasmic vesicle membrane / cell-cell junction ...Transport of glycerol from adipocytes to the liver by Aquaporins / glycerol channel activity / urea transmembrane transporter activity / Passive transport by Aquaporins / glycerol transmembrane transport / lipid droplet / cytoplasmic vesicle membrane / cell-cell junction ...Transport of glycerol from adipocytes to the liver by Aquaporins / glycerol channel activity / urea transmembrane transporter activity / Passive transport by Aquaporins / glycerol transmembrane transport /  water channel activity / water transport / water channel activity / water transport /  lipid droplet / cytoplasmic vesicle membrane / cell-cell junction / lipid droplet / cytoplasmic vesicle membrane / cell-cell junction /  cell cortex / basolateral plasma membrane / cell cortex / basolateral plasma membrane /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.55 Å cryo EM / Resolution: 2.55 Å | |||||||||
 Authors Authors | Huang P / Venskutonyte R / Fan X / Li P / Yan N / Gourdon P / Lindkvist-Petersson K | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-EM structure supports a role of AQP7 as a junction protein. Authors: Peng Huang / Raminta Venskutonytė / Rashmi B Prasad / Hamidreza Ardalani / Sofia W de Maré / Xiao Fan / Ping Li / Peter Spégel / Nieng Yan / Pontus Gourdon / Isabella Artner / Karin Lindkvist-Petersson /   Abstract: Aquaglyceroporin 7 (AQP7) facilitates glycerol flux across the plasma membrane with a critical physiological role linked to metabolism, obesity, and associated diseases. Here, we present the single- ...Aquaglyceroporin 7 (AQP7) facilitates glycerol flux across the plasma membrane with a critical physiological role linked to metabolism, obesity, and associated diseases. Here, we present the single-particle cryo-EM structure of AQP7 determined at 2.55 Å resolution adopting two adhering tetramers, stabilized by extracellularly exposed loops, in a configuration like that of the well-characterized interaction of AQP0 tetramers. The central pore, in-between the four monomers, displays well-defined densities restricted by two leucine filters. Gas chromatography mass spectrometry (GC/MS) results show that the AQP7 sample contains glycerol 3-phosphate (Gro3P), which is compatible with the identified features in the central pore. AQP7 is shown to be highly expressed in human pancreatic α- and β- cells suggesting that the identified AQP7 octamer assembly, in addition to its function as glycerol channel, may serve as junction proteins within the endocrine pancreas. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15528.map.gz emd_15528.map.gz | 125.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15528-v30.xml emd-15528-v30.xml emd-15528.xml emd-15528.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15528.png emd_15528.png | 38 KB | ||
| Others |  emd_15528_half_map_1.map.gz emd_15528_half_map_1.map.gz emd_15528_half_map_2.map.gz emd_15528_half_map_2.map.gz | 225.9 MB 225.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15528 http://ftp.pdbj.org/pub/emdb/structures/EMD-15528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15528 | HTTPS FTP |
-Related structure data
| Related structure data |  8amxMC  8amwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15528.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15528.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8464 Å | ||||||||||||||||||||||||||||||||||||
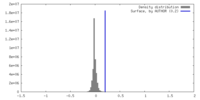
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15528_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
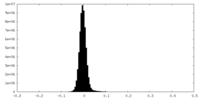
| Density Histograms |
-Half map: #1
| File | emd_15528_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
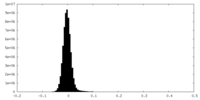
| Density Histograms |
- Sample components
Sample components
-Entire : dimer of tetramers
| Entire | Name: dimer of tetramers |
|---|---|
| Components |
|
-Supramolecule #1: dimer of tetramers
| Supramolecule | Name: dimer of tetramers / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Aquaporin-7
| Macromolecule | Name: Aquaporin-7 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.264395 KDa |
| Recombinant expression | Organism:   Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MVQASGHRRS TRGSKMVSWS VIAKIQEILQ RKMVREFLAE FMSTYVMMVF GLGSVAHMVL NKKYGSYLGV NLGFGFGVTM GVHVAGRIS GAHMNAAVTF ANCALGRVPW RKFPVYVLGQ FLGSFLAAAT IYSLFYTAIL HFSGGQLMVT GPVATAGIFA T YLPDHMTL ...String: MVQASGHRRS TRGSKMVSWS VIAKIQEILQ RKMVREFLAE FMSTYVMMVF GLGSVAHMVL NKKYGSYLGV NLGFGFGVTM GVHVAGRIS GAHMNAAVTF ANCALGRVPW RKFPVYVLGQ FLGSFLAAAT IYSLFYTAIL HFSGGQLMVT GPVATAGIFA T YLPDHMTL WRGFLNEAWL TGMLQLCLFA ITDQENNPAL PGTEALVIGI LVVIIGVSLG MNTGYAINPS RDLPPRIFTF IA GWGKQVF SNGENWWWVP VVAPLLGAYL GGIIYLVFIG STIPREPLKL EDSVAYEDHG ITVLPKMGSH EPTISPLTPV SVS PANRSS VHPAPPLHES MALEHF |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 57 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Details: 20mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK II / Details: 3s blot, 3s wait, 0s drain time, 0 blot force. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 14000 / Average exposure time: 2.0 sec. / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 371567 Details: Those particles were picked from the sub-dataset of 3225 micrographs by template picking tool. |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final 3D classification | Number classes: 5 / Software - Name: cryoSPARC |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: D4 (2x4 fold dihedral ) / Resolution.type: BY AUTHOR / Resolution: 2.55 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 102367 ) / Resolution.type: BY AUTHOR / Resolution: 2.55 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 102367 |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 98.6 |
|---|---|
| Output model |  PDB-8amx: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)