[English] 日本語
 Yorodumi
Yorodumi- EMDB-14457: Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptam... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information: / : /  HIV-1 retropepsin / HIV-1 retropepsin /  retroviral ribonuclease H / retroviral ribonuclease H /  exoribonuclease H / exoribonuclease H /  exoribonuclease H activity / host multivesicular body / DNA integration / exoribonuclease H activity / host multivesicular body / DNA integration /  RNA-directed DNA polymerase / viral genome integration into host DNA ...: / : / RNA-directed DNA polymerase / viral genome integration into host DNA ...: / : /  HIV-1 retropepsin / HIV-1 retropepsin /  retroviral ribonuclease H / retroviral ribonuclease H /  exoribonuclease H / exoribonuclease H /  exoribonuclease H activity / host multivesicular body / DNA integration / exoribonuclease H activity / host multivesicular body / DNA integration /  RNA-directed DNA polymerase / viral genome integration into host DNA / viral penetration into host nucleus / establishment of integrated proviral latency / RNA-directed DNA polymerase / viral genome integration into host DNA / viral penetration into host nucleus / establishment of integrated proviral latency /  RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity /  Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / symbiont-mediated suppression of host gene expression / viral nucleocapsid / DNA recombination / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / symbiont-mediated suppression of host gene expression / viral nucleocapsid / DNA recombination /  Hydrolases; Acting on ester bonds / Hydrolases; Acting on ester bonds /  DNA-directed DNA polymerase / aspartic-type endopeptidase activity / DNA-directed DNA polymerase / aspartic-type endopeptidase activity /  DNA-directed DNA polymerase activity / symbiont entry into host cell / DNA-directed DNA polymerase activity / symbiont entry into host cell /  lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity /  proteolysis / proteolysis /  DNA binding / DNA binding /  RNA binding / zinc ion binding / RNA binding / zinc ion binding /  membrane membraneSimilarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus type 1 BH10 / synthetic construct (others) Human immunodeficiency virus type 1 BH10 / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.32 Å cryo EM / Resolution: 3.32 Å | |||||||||
 Authors Authors | Singh AK / Das K | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine. Authors: Abhimanyu K Singh / Brent De Wijngaert / Marc Bijnens / Kris Uyttersprot / Hoai Nguyen / Sergio E Martinez / Dominique Schols / Piet Herdewijn / Christophe Pannecouque / Eddy Arnold / Kalyan Das /   Abstract: Structures trapping a variety of functional and conformational states of HIV-1 reverse transcriptase (RT) have been determined by X-ray crystallography. These structures have played important roles ...Structures trapping a variety of functional and conformational states of HIV-1 reverse transcriptase (RT) have been determined by X-ray crystallography. These structures have played important roles in explaining the mechanisms of catalysis, inhibition, and drug resistance and in driving drug design. However, structures of several desired complexes of RT could not be obtained even after many crystallization or crystal soaking experiments. The ternary complexes of doravirine and rilpivirine with RT/DNA are such examples. Structural study of HIV-1 RT by single-particle cryo-electron microscopy (cryo-EM) has been challenging due to the enzyme's relatively smaller size and higher flexibility. We optimized a protocol for rapid structure determination of RT complexes by cryo-EM and determined six structures of wild-type and E138K/M184I mutant RT/DNA in complexes with the nonnucleoside inhibitors rilpivirine, doravirine, and nevirapine. RT/DNA/rilpivirine and RT/DNA/doravirine complexes have structural differences between them and differ from the typical conformation of nonnucleoside RT inhibitor (NNRTI)-bound RT/double-stranded DNA (dsDNA), RT/RNA-DNA, and RT/dsRNA complexes; the primer grip in RT/DNA/doravirine and the YMDD motif in RT/DNA/rilpivirine have large shifts. The DNA primer 3'-end in the doravirine-bound structure is positioned at the active site, but the complex is in a nonproductive state. In the mutant RT/DNA/rilpivirine structure, I184 is stacked with the DNA such that their relative positioning can influence rilpivirine in the pocket. Simultaneously, E138K mutation opens the NNRTI-binding pocket entrance, potentially contributing to a faster rate of rilpivirine dissociation by E138K/M184I mutant RT, as reported by an earlier kinetic study. These structural differences have implications for understanding molecular mechanisms of drug resistance and for drug design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14457.map.gz emd_14457.map.gz | 6.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14457-v30.xml emd-14457-v30.xml emd-14457.xml emd-14457.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14457_fsc.xml emd_14457_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_14457.png emd_14457.png | 77.8 KB | ||
| Others |  emd_14457_half_map_1.map.gz emd_14457_half_map_1.map.gz emd_14457_half_map_2.map.gz emd_14457_half_map_2.map.gz | 6.9 MB 6.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14457 http://ftp.pdbj.org/pub/emdb/structures/EMD-14457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14457 | HTTPS FTP |
-Related structure data
| Related structure data |  7z24MC  7z29C  7z2dC  7z2eC  7z2gC  7z2hC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14457.map.gz / Format: CCP4 / Size: 7.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14457.map.gz / Format: CCP4 / Size: 7.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_14457_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_14457_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 reverse transcriptase with a DNA aptamer in complex with ne...
| Entire | Name: HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 reverse transcriptase with a DNA aptamer in complex with ne...
| Supramolecule | Name: HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus type 1 BH10 Human immunodeficiency virus type 1 BH10 |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
-Supramolecule #2: HIV-1 REVERSE TRANSCRIPTASE P66 SUBUNIT
| Supramolecule | Name: HIV-1 REVERSE TRANSCRIPTASE P66 SUBUNIT / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus type 1 BH10 Human immunodeficiency virus type 1 BH10 |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
-Supramolecule #3: HIV-1 REVERSE TRANSCRIPTASE P51 SUBUNIT
| Supramolecule | Name: HIV-1 REVERSE TRANSCRIPTASE P51 SUBUNIT / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus type 1 BH10 Human immunodeficiency virus type 1 BH10 |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
-Macromolecule #1: Reverse transcriptase/ribonuclease H
| Macromolecule | Name: Reverse transcriptase/ribonuclease H / type: protein_or_peptide / ID: 1 / Details: P66 SUBUNIT / Number of copies: 1 / Enantiomer: LEVO / EC number:  RNA-directed DNA polymerase RNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus type 1 BH10 / Strain: isolate BH10 Human immunodeficiency virus type 1 BH10 / Strain: isolate BH10 |
| Molecular weight | Theoretical: 64.037383 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MVPISPIETV PVKLKPGMDG PKVKQWPLTE EKIKALVEIC TEMEKEGKIS KIGPENPYNT PVFACKKKDS TKWRKLVDFR ELNKRTQDF WEVQLGIPHP AGLKKKKSVT VLDVGDAYFS VPLDEDFRKY TAFTIPSINN ETPGIRYQYN VLPQGWKGSP A IFQSSMTK ...String: MVPISPIETV PVKLKPGMDG PKVKQWPLTE EKIKALVEIC TEMEKEGKIS KIGPENPYNT PVFACKKKDS TKWRKLVDFR ELNKRTQDF WEVQLGIPHP AGLKKKKSVT VLDVGDAYFS VPLDEDFRKY TAFTIPSINN ETPGIRYQYN VLPQGWKGSP A IFQSSMTK ILEPFKKQNP DIVIYQYMDD LYVGSDLEIG QHRTKIEELR QHLLRWGLTT PDKKHQKEPP FLWMGYELHP DK WTVQPIV LPEKDSWTVN DIQKLVGKLN WASQIYPGIK VRQLSKLLRG TKALTEVIPL TEEAELELAE NREILKEPVH GVY YDPSKD LIAEIQKQGQ GQWTYQIYQE PFKNLKTGKY ARMRGAHTND VKQLTEAVQK ITTESIVIWG KTPKFKLPIQ KETW ETWWT EYWQATWIPE WEFVNTPPLV KLWYQLEKEP IVGAETFYVD GAANRETKLG KAGYVTNKGR QKVVPLTNTT NQKTE LQAI YLALQDSGLE VNIVTNSQYA LGIIQAQPDK SESELVNQII EQLIKKEKVY LAWVPAHKGI GGNEQVDKLV SA |
-Macromolecule #2: Reverse transcriptase/ribonuclease H
| Macromolecule | Name: Reverse transcriptase/ribonuclease H / type: protein_or_peptide / ID: 2 / Details: P51 subunit / Number of copies: 1 / Enantiomer: LEVO / EC number:  RNA-directed DNA polymerase RNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus type 1 BH10 Human immunodeficiency virus type 1 BH10 |
| Molecular weight | Theoretical: 50.039488 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: PISPIETVPV KLKPGMDGPK VKQWPLTEEK IKALVEICTE MEKEGKISKI GPENPYNTPV FAIKKKDSTK WRKLVDFREL NKRTQDFWE VQLGIPHPAG LKKKKSVTVL DVGDAYFSVP LDEDFRKYTA FTIPSINNET PGIRYQYNVL PQGWKGSPAI F QSSMTKIL ...String: PISPIETVPV KLKPGMDGPK VKQWPLTEEK IKALVEICTE MEKEGKISKI GPENPYNTPV FAIKKKDSTK WRKLVDFREL NKRTQDFWE VQLGIPHPAG LKKKKSVTVL DVGDAYFSVP LDEDFRKYTA FTIPSINNET PGIRYQYNVL PQGWKGSPAI F QSSMTKIL EPFKKQNPDI VIYQYMDDLY VGSDLEIGQH RTKIEELRQH LLRWGLTTPD KKHQKEPPFL WMGYELHPDK WT VQPIVLP EKDSWTVNDI QKLVGKLNWA SQIYPGIKVR QLSKLLRGTK ALTEVIPLTE EAELELAENR EILKEPVHGV YYD PSKDLI AEIQKQGQGQ WTYQIYQEPF KNLKTGKYAR MRGAHTNDVK QLTEAVQKIT TESIVIWGKT PKFKLPIQKE TWET WWTEY WQATWIPEWE FVNTPPLVKL WYQ |
-Macromolecule #3: DNA (38-mer)
| Macromolecule | Name: DNA (38-mer) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 11.739513 KDa |
| Sequence | String: (DT)(DA)(DA)(DT)(DT)(DC)(OMC)(DC)(OMC)(DC) (DC)(DC)(DT)(DT)(DC)(DG)(DG)(DT)(DG) (DC)(DT)(DT)(DT)(DG)(DC)(DA)(DC)(DC)(DG) (DA)(DA)(DG)(DG)(DG)(DG)(DG)(DG)(DG) |
-Macromolecule #4: 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1...
| Macromolecule | Name: 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE type: ligand / ID: 4 / Number of copies: 1 / Formula: NVP |
|---|---|
| Molecular weight | Theoretical: 266.298 Da |
| Chemical component information | 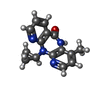 ChemComp-NVP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.03 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 281 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 150000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 150000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1910 / Average exposure time: 55.0 sec. / Average electron dose: 40.0 e/Å2 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






































