[English] 日本語
 Yorodumi
Yorodumi- EMDB-1356: Structural basis for the PufX-mediated dimerization of bacterial ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1356 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis for the PufX-mediated dimerization of bacterial photosynthetic core complexes. | |||||||||
 Map data Map data | 3D map file of Rhobacter veldkampii LH1-RC obtained by cryoEM and low-pass filtered at a resolution of 11 angstroems | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Rhodobacter veldkampii (bacteria) Rhodobacter veldkampii (bacteria) | |||||||||
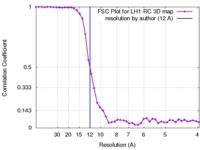
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / cryo EM /  negative staining / Resolution: 12.0 Å negative staining / Resolution: 12.0 Å | |||||||||
 Authors Authors | Busselez J / Cottevieille M / Cuniasse P / Boisset N / Levy D | |||||||||
 Citation Citation |  Journal: Structure / Year: 2007 Journal: Structure / Year: 2007Title: Structural basis for the PufX-mediated dimerization of bacterial photosynthetic core complexes. Authors: Johan Busselez / Magali Cottevieille / Philippe Cuniasse / Francesca Gubellini / Nicolas Boisset / Daniel Lévy /  Abstract: In Rhodobacter (Rba.) sphaeroides, the subunit PufX is involved in the dimeric organization of the core complex. Here, we report the 3D reconstruction at 12 A by cryoelectron microscopy of the core ...In Rhodobacter (Rba.) sphaeroides, the subunit PufX is involved in the dimeric organization of the core complex. Here, we report the 3D reconstruction at 12 A by cryoelectron microscopy of the core complex of Rba. veldkampii, a complex of approximately 300 kDa without symmetry. The core complex is monomeric and constituted by a light-harvesting complex 1 (LH1) ring surrounding a uniquely oriented reaction center (RC). The LH1 consists of 15 resolved alpha/beta heterodimers and is interrupted. Within the opening, PufX polypeptide is assigned at a position facing the Q(B) site of the RC. This core complex is different from a dissociated dimer of the core complex of Rba. sphaeroides revealing that PufX in Rba. veldkampii is unable to dimerize. The absence in PufX of Rba. veldkampii of a G(31)XXXG(35) dimerization motif highlights the transmembrane interactions between PufX subunits involved in the dimerization of the core complexes of Rhodobacter species. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1356.map.gz emd_1356.map.gz | 5.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1356-v30.xml emd-1356-v30.xml emd-1356.xml emd-1356.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_1356_fsc.xml emd_1356_fsc.xml | 4.3 KB | Display |  FSC data file FSC data file |
| Images |  EMD-1356.png EMD-1356.png | 148.4 KB | ||
| Others |  FSC_graph_emd_1356.tif FSC_graph_emd_1356.tif | 145.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1356 http://ftp.pdbj.org/pub/emdb/structures/EMD-1356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1356 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1356.map.gz / Format: CCP4 / Size: 5.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1356.map.gz / Format: CCP4 / Size: 5.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map file of Rhobacter veldkampii LH1-RC obtained by cryoEM and low-pass filtered at a resolution of 11 angstroems | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.95 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Others
- Sample components
Sample components
-Entire : Core complex of Rhodobacter veldkampii
| Entire | Name: Core complex of Rhodobacter veldkampii |
|---|---|
| Components |
|
-Supramolecule #1000: Core complex of Rhodobacter veldkampii
| Supramolecule | Name: Core complex of Rhodobacter veldkampii / type: sample / ID: 1000 / Oligomeric state: monomers / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: core complex of Rba. veldkampii
| Macromolecule | Name: core complex of Rba. veldkampii / type: protein_or_peptide / ID: 1 / Name.synonym: LH1-RC of Rba. veldkampii / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Rhodobacter veldkampii (bacteria) / Strain: DSM 11550 Rhodobacter veldkampii (bacteria) / Strain: DSM 11550 |
| Molecular weight | Experimental: 300 KDa |
-Experimental details
-Structure determination
| Method |  negative staining, negative staining,  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: Glycine-glycine 50 mM, NaCl 200 mM, DOTM 0.1 % |
| Staining | Type: NEGATIVE Details: CRYOEM : 4 microL were applied on a Lacey Formwar grid. |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 93 K / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: manual plunger / Method: Manual single-sided blotting |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 45000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 3.46 µm / Nominal defocus min: 1.87 µm / Nominal magnification: 45000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 3.46 µm / Nominal defocus min: 1.87 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder: Gatan / Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Min: 93 K / Max: 94 K |
| Details | low-dose illumination |
| Date | Jun 1, 2005 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Number real images: 74 / Average electron dose: 10 e/Å2 / Details: Scanner model : Nikon Coolscan 8000ED / Bits/pixel: 8 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)